Counting empty circles
|
Hello,
I have images (see below) that have many glandular trichomes (the round shiny objects) which I would like to quantify. I simply want to know how many of them there are. It seems like I should be able to ask ImageJ to count the number of these glandular trichomes, but I don't know how to do this. I have tried many things already, but have hit a wall. The smaller circles within each gland seem to be very consistent in size. I wonder if this consistency in size can be used somehow. Thanks so much! Sincerely, Scott Chamberlain Rice University, EEB Dept. 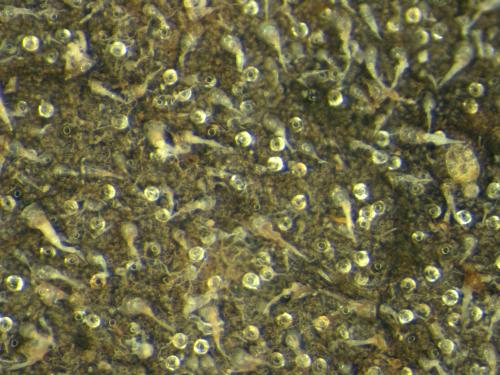
|
|
Dear Scott,
You have a very interesting image processing challenge here. What you are suggesting to do, is to segment the image by a form of pattern recognition: recognizing particles with a certain size of hole. Not the easiest of image processing tasks in general, but because you particles are more or less circular (and hence rotation invariant), you might just have one of those cases where it is not totally impossible. I tried calculating the crosscorrelation between the image you refer to and another image that contained a white 'dougnut' with the approximate size of a trichome. If all is well, this should give an image where the highest pixel values correspond to the best template matches. By using "process>find maxima..." you can select them and count them. Bad new though, it didn't work, I'm sorry to say. Most matches did not correspond to trichomes and I didn't have time to do a proper analysis about why this was. I'm confident though, that with some additional tweaking it still might work, but I can guarantee that you that there will always be a larger error margin on the eventual count of trichomes. Unfortunately, template matching is something that we humans just do much,much better than any computer can even dream of... Good luck, Kind regards, Jaap Kokorian 2011/1/4 schamber789 <[hidden email]>: > Hello, > > I have images (see below) that have many glandular trichomes (the round > shiny objects) which I would like to quantify. I simply want to know how > many of them there are. It seems like I should be able to ask ImageJ to > count the number of these glandular trichomes, but I don't know how to do > this. I have tried many things already, but have hit a wall. The smaller > circles within each gland seem to be very consistent in size. I wonder if > this consistency in size can be used somehow. Thanks so much! > > Sincerely, > > Scott Chamberlain > Rice University, EEB Dept. > > > http://imagej.588099.n2.nabble.com/file/n5889708/2309-1.jpg > -- > View this message in context: http://imagej.588099.n2.nabble.com/Counting-empty-circles-tp5889708p5889708.html > Sent from the ImageJ mailing list archive at Nabble.com. > |
|
You may find that it is much faster to take advantage of the " template matching is something that we humans just do
much,much better than any computer can even dream of..." and use the Cell Counter. If there are too many of these glands, you may subsample in a uniform random fashion and get away with counting only about 200 per specimen. http://www.stereology.info/counting-rules/ Anda Cornea, Ph.D. Director of the Imaging Core Oregon National Primate Research Center Oregon Heath & Science University 503-690-5293 -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Jaap Kokorian Sent: Wednesday, January 05, 2011 4:55 AM To: [hidden email] Subject: Re: Counting empty circles Dear Scott, You have a very interesting image processing challenge here. What you are suggesting to do, is to segment the image by a form of pattern recognition: recognizing particles with a certain size of hole. Not the easiest of image processing tasks in general, but because you particles are more or less circular (and hence rotation invariant), you might just have one of those cases where it is not totally impossible. I tried calculating the crosscorrelation between the image you refer to and another image that contained a white 'dougnut' with the approximate size of a trichome. If all is well, this should give an image where the highest pixel values correspond to the best template matches. By using "process>find maxima..." you can select them and count them. Bad new though, it didn't work, I'm sorry to say. Most matches did not correspond to trichomes and I didn't have time to do a proper analysis about why this was. I'm confident though, that with some additional tweaking it still might work, but I can guarantee that you that there will always be a larger error margin on the eventual count of trichomes. Unfortunately, template matching is something that we humans just do much,much better than any computer can even dream of... Good luck, Kind regards, Jaap Kokorian 2011/1/4 schamber789 <[hidden email]>: > Hello, > > I have images (see below) that have many glandular trichomes (the round > shiny objects) which I would like to quantify. I simply want to know how > many of them there are. It seems like I should be able to ask ImageJ to > count the number of these glandular trichomes, but I don't know how to do > this. I have tried many things already, but have hit a wall. The smaller > circles within each gland seem to be very consistent in size. I wonder if > this consistency in size can be used somehow. Thanks so much! > > Sincerely, > > Scott Chamberlain > Rice University, EEB Dept. > > > http://imagej.588099.n2.nabble.com/file/n5889708/2309-1.jpg > -- > View this message in context: http://imagej.588099.n2.nabble.com/Counting-empty-circles-tp5889708p5889708.html > Sent from the ImageJ mailing list archive at Nabble.com. > |
|
Thanks very much for all of your comments regarding my question. I still have not found a method that works, but am still working on it.
I agree that it is technically easiest to simply count the trichomes manually. However, counting manually takes a lot of time, which is equivalent to a lot of money to pay a student to do the counting. Unfortunately, as a graduate student working on research independent from their advisor, there isn't a lot of money to be had. Thus, finding a technological solution is superior as it saves money and is faster, once the solution is found. To your subsampling comment Anda, the leaf discs are already subsamples of whole leaves, so I don't want to do further subsampling which would reduce my confidence in our estimate of leaf trichome abundance/density. Thanks again, Scott On Jan 5, 2011, at 12:32 PM, Anda Cornea wrote: > You may find that it is much faster to take advantage of the " template matching is something that we humans just do > much,much better than any computer can even dream of..." and use the Cell Counter. > > If there are too many of these glands, you may subsample in a uniform random fashion and get away with counting only about 200 per specimen. http://www.stereology.info/counting-rules/ > > > Anda Cornea, Ph.D. > Director of the Imaging Core > Oregon National Primate Research Center > Oregon Heath & Science University > 503-690-5293 > > -----Original Message----- > From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Jaap Kokorian > Sent: Wednesday, January 05, 2011 4:55 AM > To: [hidden email] > Subject: Re: Counting empty circles > > Dear Scott, > > You have a very interesting image processing challenge here. What you > are suggesting to do, is to segment the image by a form of pattern > recognition: recognizing particles with a certain size of hole. Not > the easiest of image processing tasks in general, but because you > particles are more or less circular (and hence rotation invariant), > you might just have one of those cases where it is not totally > impossible. > > I tried calculating the crosscorrelation between the image you refer > to and another image that contained a white 'dougnut' with the > approximate size of a trichome. If all is well, this should give an > image where the highest pixel values correspond to the best template > matches. By using "process>find maxima..." you can select them and > count them. > > Bad new though, it didn't work, I'm sorry to say. Most matches did not > correspond to trichomes and I didn't have time to do a proper analysis > about why this was. I'm confident though, that with some additional > tweaking it still might work, but I can guarantee that you that there > will always be a larger error margin on the eventual count of > trichomes. > > Unfortunately, template matching is something that we humans just do > much,much better than any computer can even dream of... > > Good luck, > Kind regards, > > Jaap Kokorian > > > > 2011/1/4 schamber789 <[hidden email]>: >> Hello, >> >> I have images (see below) that have many glandular trichomes (the round >> shiny objects) which I would like to quantify. I simply want to know how >> many of them there are. It seems like I should be able to ask ImageJ to >> count the number of these glandular trichomes, but I don't know how to do >> this. I have tried many things already, but have hit a wall. The smaller >> circles within each gland seem to be very consistent in size. I wonder if >> this consistency in size can be used somehow. Thanks so much! >> >> Sincerely, >> >> Scott Chamberlain >> Rice University, EEB Dept. >> >> >> http://imagej.588099.n2.nabble.com/file/n5889708/2309-1.jpg >> -- >> View this message in context: http://imagej.588099.n2.nabble.com/Counting-empty-circles-tp5889708p5889708.html >> Sent from the ImageJ mailing list archive at Nabble.com. >> |
|
2011/1/6 Scott Chamberlain <[hidden email]>:
> Thanks very much for all of your comments regarding my question. I still have not found a method that works, but am still working on it. > > I agree that it is technically easiest to simply count the trichomes manually. However, counting manually takes a lot of time, which is equivalent to a lot of money to pay a student to do the counting. Unfortunately, as a graduate student working on research independent from their advisor, there isn't a lot of money to be had. Thus, finding a technological solution is superior as it saves money and is faster, once the solution is found. > > To your subsampling comment Anda, the leaf discs are already subsamples of whole leaves, so I don't want to do further subsampling which would reduce my confidence in our estimate of leaf trichome abundance/density. > > Thanks again, Scott Scott, Have a look at these two methods: SIOX: http://pacific.mpi-cbg.de/wiki/index.php/SIOX:_Simple_Interactive_Object_Extraction Trainable Segmentation: http://pacific.mpi-cbg.de/wiki/index.php/Trainable_Segmentation_Plugin I gave it a go at the second and I managed to get a reasonable mask. Then I dilated, closed holes, run watershed, and run analyze particles to count them. The count was off by 1 too many. Albert -- http://albert.rierol.net |
|
Hi Albert, Thanks very much!
I tried that second method, Trainable Segmentation, in Fiji, but I could not get good results as you did. I assumed by second, you mean the second URL? Can you send an image of what objects you delineated to train the program? Best regards, Scott Chamberlain On Jan 6, 2011, at 10:06 AM, Albert Cardona wrote: > 2011/1/6 Scott Chamberlain <[hidden email]>: >> Thanks very much for all of your comments regarding my question. I still have not found a method that works, but am still working on it. >> >> I agree that it is technically easiest to simply count the trichomes manually. However, counting manually takes a lot of time, which is equivalent to a lot of money to pay a student to do the counting. Unfortunately, as a graduate student working on research independent from their advisor, there isn't a lot of money to be had. Thus, finding a technological solution is superior as it saves money and is faster, once the solution is found. >> >> To your subsampling comment Anda, the leaf discs are already subsamples of whole leaves, so I don't want to do further subsampling which would reduce my confidence in our estimate of leaf trichome abundance/density. >> >> Thanks again, Scott > > > Scott, > > Have a look at these two methods: > > SIOX: > http://pacific.mpi-cbg.de/wiki/index.php/SIOX:_Simple_Interactive_Object_Extraction > > Trainable Segmentation: > http://pacific.mpi-cbg.de/wiki/index.php/Trainable_Segmentation_Plugin > > I gave it a go at the second and I managed to get a reasonable mask. > Then I dilated, closed holes, run watershed, and run analyze particles > to count them. The count was off by 1 too many. > > Albert > > -- > http://albert.rierol.net |
|
2011/1/6 Scott Chamberlain <[hidden email]>:
> Hi Albert, Thanks very much! > > I tried that second method, Trainable Segmentation, in Fiji, but I could not get good results as you did. I assumed by second, you mean the second URL? Can you send an image of what objects you delineated to train the program? I've attached a screenshot. What improves training lots is to use more feature descriptors. Notice all checkboxes are selected in the "Settings" dialog. I've also increased features to "4", but the improvement is almost not worth it. The second filtering is done by size in the "Analyze particles". I used "100-infinity", after measuring individual masks and finding that they should be around 200 px in area. After spending more than 30 s on the image, I see there's more error than just 1 off. Perhaps you could count in a few images manually and find out if the error is consistent, then estimate a correction factor. Albert -- http://albert.rierol.net |
|
In reply to this post by schamber789
Hi Scott,
you could try my 'Feature Finder' template matching plugin - I have put it online now: http://imagejdocu.tudor.lu/doku.php? id=plugin:analysis:feature_finder:start It seems that the features of interest (trichomes) can appear differently, with or without the bright ring? So you might combine the results obtained with different templates (prototypes). And there are lots of parameters to try: different brightness/ contrast of the template, different ROI size in the template, and all the parameters of the dialog box... Michael ________________________________________________________________ > Hello, > > I have images (see below) that have many glandular trichomes (the > round > shiny objects) which I would like to quantify. I simply want to > know how > many of them there are. It seems like I should be able to ask > ImageJ to > count the number of these glandular trichomes, but I don't know how > to do > this. I have tried many things already, but have hit a wall. The > smaller > circles within each gland seem to be very consistent in size. I > wonder if > this consistency in size can be used somehow. Thanks so much! > > Sincerely, > > Scott Chamberlain > Rice University, EEB Dept. |
|
Hi Michael,
I wonder if Feature Finder plugin can be automated somehow. I know the website says that it does not process stacks, but is there any other way to process many images somehow with Feature Finder? I have thousands to process. Thanks! Scott On Jan 7, 2011, at 5:21 AM, Michael Schmid wrote: > Hi Scott, > > you could try my 'Feature Finder' template matching plugin - I have put it online now: > http://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:feature_finder:start > > It seems that the features of interest (trichomes) can appear differently, with or without the bright ring? So you might combine the results obtained with different templates (prototypes). > And there are lots of parameters to try: different brightness/contrast of the template, different ROI size in the template, and all the parameters of the dialog box... > > Michael > ________________________________________________________________ > >> Hello, >> >> I have images (see below) that have many glandular trichomes (the round >> shiny objects) which I would like to quantify. I simply want to know how >> many of them there are. It seems like I should be able to ask ImageJ to >> count the number of these glandular trichomes, but I don't know how to do >> this. I have tried many things already, but have hit a wall. The smaller >> circles within each gland seem to be very consistent in size. I wonder if >> this consistency in size can be used somehow. Thanks so much! >> >> Sincerely, >> >> Scott Chamberlain >> Rice University, EEB Dept. > <Feature_Finder_Test.jpg> |
|
Hi Scott,
with the exception of the 'refine' button, the Feature Finder plugin can be called in a macro. You can record the syntax with Plugins>Macros>Record. n = nSlices(); for (i=1; i<=n; i++) { run ("Feature Finder", FeatureFinderOptions); } In case you want a more elaborate function than just 'count' you may also have a look at the FindStackMaxima macro for an example http://rsb.info.nih.gov/ij/macros/FindStackMaxima.txt (by the way, in the meanwhile the FindStackMaxima macro as such is obsolete because Find Maxima can process stacks) Michael ________________________________________________________________ On Fri, January 7, 2011 21:01, Scott Chamberlain wrote: > Hi Michael, > > I wonder if Feature Finder plugin can be automated somehow. I know the > website says that it does not process stacks, but is there any other way > to process many images somehow with Feature Finder? I have thousands to > process. > > Thanks! Scott > > On Jan 7, 2011, at 5:21 AM, Michael Schmid wrote: > >> Hi Scott, >> >> you could try my 'Feature Finder' template matching plugin - I have put >> it online now: >> http://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:feature_finder:start >> >> It seems that the features of interest (trichomes) can appear >> differently, with or without the bright ring? So you might combine the >> results obtained with different templates (prototypes). >> And there are lots of parameters to try: different brightness/contrast >> of the template, different ROI size in the template, and all the >> parameters of the dialog box... >> >> Michael >> ________________________________________________________________ >> >>> Hello, >>> >>> I have images (see below) that have many glandular trichomes (the round >>> shiny objects) which I would like to quantify. I simply want to know >>> how >>> many of them there are. It seems like I should be able to ask ImageJ >>> to >>> count the number of these glandular trichomes, but I don't know how to >>> do >>> this. I have tried many things already, but have hit a wall. The >>> smaller >>> circles within each gland seem to be very consistent in size. I wonder >>> if >>> this consistency in size can be used somehow. Thanks so much! >>> >>> Sincerely, >>> >>> Scott Chamberlain >>> Rice University, EEB Dept. >> <Feature_Finder_Test.jpg> > |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

