Cross section area of muscle, less the fat
|
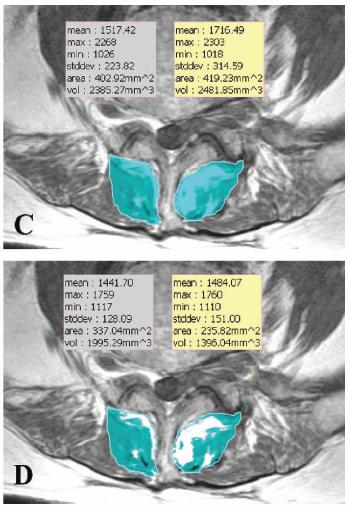
On MRI images, I am trying to measure the cross sectional area of muscle after subtracting out the fat component. I figured out one way to do it with ImageJ, but it is very labor intensive. Basically, I draw a region of interest, make a histogram of the pixel intensities, cut/paste the histogram list into excel, manually choose a pixel intensity cut-off between fat and muscle, count pixels on "muscle side" of cut-off, and then convert the number of pixels into an area measurement. Does anyone know of a faster or automated way to do this? Perhaps there is a different software program that is free or inexpensive that can do this? The attached image is an example of what I would like to do, performed with a Korean program called Rapidia. Unfortunately, that program is quite expensive. Any ideas? Thanks
|
Re: Cross section area of muscle, less the fat
|
Using Image->Adjust->Threshold, you can achieve the same result as
pasting the histogram into Excel and choosing a cutoff. Note that threshold only works on 8-bit grayscale images, and when thresholding the histogram displayed in the dialog is for the ROI if one is selected (otherwise it's the histogram of the whole image). In Analyze->Set Scale, you can set a conversion factor to change pixels to some real world unit. You will need a scale mark in the image or some knowledge of your imaging setup; a scale mark is the easiest solution. In Analyze->Set Measurements you can choose the results you want to measure. You should have "Limit to Threshold" selected, and when you choose your threshold, make sure that the portion you want to measure is red. Muscle tissue in your case. When you have set the scale and the measurements you want, you can select your ROI, pick the threshold, then use Analyze->Measure, and the results will be shown in the results table. If you want to have multiple ROI, such as in the Rapidia image you linked, you can try the ROI manager (Analyze->Tools->ROI manager). ImageJ will list the results in the results table (which can be saved), but if you wanted you could make a macro to draw them onto the image like Rapidia does. I like my results in text files though since they are easier to deal with later. That should speed things up a bit. If the scale doesn't change between images, you can leave it alone and process several images at a time. As far as more automation, if your images are like the one you linked, then you will almost certainly need to draw the ROI by hand. However, you may be able to use an autothreshold instead of choosing one manually. ImageJ's threshold dialog has an auto button that might work, but if you check the plug-in site, there are thresholders that use different algorithms. Also you can read about the macro language (http://rsbweb.nih.gov/ij/developer/macro/macros.html) which can speed up repetitive tasks, like naming ROI. Justin On Sat, Dec 13, 2008 at 1:22 PM, casab <[hidden email]> wrote: > On MRI images, I am trying to measure the cross sectional area of muscle > after subtracting out the fat component. I figured out one way to do it > with ImageJ, but it is very labor intensive. Basically, I draw a region of > interest, make a histogram of the pixel intensities, cut/paste the histogram > list into excel, manually choose a pixel intensity cut-off between fat and > muscle, count pixels on "muscle side" of cut-off, and then convert the > number of pixels into an area measurement. Does anyone know of a faster or > automated way to do this? Perhaps there is a different software program > that is free or inexpensive that can do this? The attached image is an > example of what I would like to do, performed with a Korean program called > Rapidia. Unfortunately, that program is quite expensive. Any ideas? > Thanks > > > http://n2.nabble.com/file/n1652546/multifidus.jpg > > -- > View this message in context: http://n2.nabble.com/Cross-section-area-of-muscle%2C-less-the-fat-tp1652546p1652546.html > Sent from the ImageJ mailing list archive at Nabble.com. > |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

