Deconvolution Lab "Batch" Module
12




12
|
Hi,
Has anyone tried the "Batch" module in deconvolution lab with ImageJ ? I have tried, but I cannot input multiple data sets, I don't know why. When I select to input a folder, it told me there's no data input. But if I select the tiff images, it says my input PSF has greater dimention than images. Is there anyone can help? Thanks, Mirrownight |
|
Hi mirrownight
Is there any possibility that the PSF is 3D and the images are 2D?? Maybe that is what the message "PSF has greater dimension than images" means. Are you able to share the data?? If so I can try and run it. Brian On Wed, Mar 2, 2016 at 9:37 PM, mirrownight <[hidden email]> wrote: > Hi, > > Has anyone tried the "Batch" module in deconvolution lab with ImageJ ? > I have tried, but I cannot input multiple data sets, I don't know why. > > When I select to input a folder, it told me there's no data input. > But if I select the tiff images, it says my input PSF has greater dimention > than images. > Is there anyone can help? > > Thanks, > > Mirrownight > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Module-tp5015776.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by mirrownight
Hi Morrownight
I just did a very quick test using Deconvolution Lab with Richardson Lucy and batch mode. My images were all .tifs, 8-bit, 2 dimensions (x,y), the PSF was smaller than the image. Everything worked fine. Here are some suggestions for trouble shooting... 1. Check to make sure images and psf are indeed the same dimensions (no extra channels, or z) 2. Try making the images all .tif format 2. Try making images and PSF the same pixel type (all 8-bit, all 32 bit). On Wed, Mar 2, 2016 at 9:37 PM, mirrownight <[hidden email]> wrote: > Hi, > > Has anyone tried the "Batch" module in deconvolution lab with ImageJ ? > I have tried, but I cannot input multiple data sets, I don't know why. > > When I select to input a folder, it told me there's no data input. > But if I select the tiff images, it says my input PSF has greater dimention > than images. > Is there anyone can help? > > Thanks, > > Mirrownight > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Module-tp5015776.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Deconvolution Lab "Batch" Module
|
Hi, Brian
Thanks for your reply. I've checked both the PSF and the images are all seperate image stacks each composed of 41 slices of 2D tiff images. Hence, they have x-y dimension and z stack. Are the 3D image stacks not allowed to run Batch module in DeconvolutionLab ? I make the PSF and the images the same x-y dimension (256x256 pixel), but it still doesn't work. The link bellow is the data for test: https://drive.google.com/file/d/0BzOF8t32YFytbEszdGJ3ZkZ2RUk/view?usp=sharing Best regards, Mirrownight |
Re: Deconvolution Lab "Batch" Module
|
This post was updated on .
Hi, Brian
Here are the info of the PSF and images. 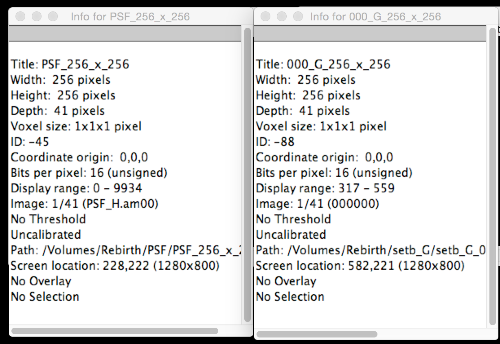 Thank you. Mirrownight |
Re: Deconvolution Lab "Batch" Module
|
Here is the link of detailed description of my question.
https://drive.google.com/file/d/0BzOF8t32YFytQVBHbmh6dU1oaTg/view?usp=sharing Thanks very much for the help. Mirrownight |
|
Hi Mirrownight
I think that deconvolutionlab does not recognize the series of tif images as a stack. I got it working by doing the following 1. Open each series as a stack by using file->import->image sequence 2. I then resaved each stack as one 3D tif file, I saved them all in the same directory. 3. I then had a new directory, with 3 files, each representing a 3D stack 4. Then I ran deconvolutionlab, and setting batch->'Input Folder' to point to the new directory. Let me know if the same process works for you On Fri, Mar 4, 2016 at 1:43 AM, mirrownight <[hidden email]> wrote: > Here is the link of detailed description of my question. > > > https://drive.google.com/file/d/0BzOF8t32YFytQVBHbmh6dU1oaTg/view?usp=sharing > < > https://drive.google.com/file/d/0BzOF8t32YFytQVBHbmh6dU1oaTg/view?usp=sharing > > > > Thanks very much for the help. > > > Mirrownight > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Module-tp5015776p5015797.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Deconvolution Lab "Batch" Module
|
Hi Brian
Thank you very, very, very much for the solution. It's wierd for me that the PSF is still 2D tiff, but the input files should be 3D tiff for recognization. I can not think of this idea.  But it's working now~ But it's working now~
Thanks again for the help!!! Mirrownight |
|
Hi Morrownight
Actually when I ran it both the PSF and Image were 3D - I loaded the PSF as a series using the ImageJ import->sequence option, so all the 2D slices were combined into a stack. - DeconvolutionLab in batch mode is not able to detect the sequence... itjust loads each file separately, thus they need to be combined into a stack before hand. On Sat, Mar 5, 2016 at 10:57 PM, mirrownight <[hidden email]> wrote: > Hi Brian > > Thank you very, very, very much for the solution. > > It's wierd for me that the PSF is still 2D tiff, but the input files should > be 3D tiff for recognization. > I can not think of this idea. But it's working now~ > > > Thanks again for the help!!! > > > Mirrownight > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Module-tp5015776p5015809.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
my input image size is 2048*2048*37 and psf size is 512*512*99....how should i resize it?do i have to do fft or ifft to get deconvolution result..the psf file that i have contain several beads....should i take average of all the beads in that file to deconvolution.... or just uploading the psf file with sveral beads is fine?
i am doing with deconvolution lab software...how should i proceed? |
|
Hi Ash
If you can share your image and psf I can try and help you. From your description I suspect your image contains multiple beads. To use it as a PSF you would have to crop out one bead. Brian On Mon, May 15, 2017 at 5:45 PM, Ash <[hidden email]> wrote: > my input image size is 2048*2048*37 and psf size is 512*512*99....how > should > i resize it?do i have to do fft or ifft to get deconvolution result..the > psf > file that i have contain several beads....should i take average of all the > beads in that file to deconvolution.... or just uploading the psf file with > sveral beads is fine? > i am doing with deconvolution lab software...how should i proceed? > > > > -- > View this message in context: http://imagej.1557.x6.nabble. > com/Deconvolution-Lab-Batch-Module-tp5015776p5018724.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
hii thank you for replying me..yes..i do have multiple bead...the psf has 99 frames and image has 37 frames....if i am cropping a single bead out then how should i adjust the size to make it compatible with the raw image???because raw images size is 2048*2048 and bead size is 512......if i crop out a single bead, then the bead file will become more smaller!!! On Tue, May 23, 2017 at 8:00 PM, bnorthan [via ImageJ] <[hidden email]> wrote: Hi Ash |
|
Hi Ash
If you are using deconvolution lab, it should automatically resize the PSF in x and y. If I recall correctly deconvolution lab does not resize in z. So you should make the PSF x by y by 37 (z same z in image). Make x and y the right size to contain one bead. Make sure the bead is centered. I also would subtract the background level (at least subtract the min value of the cropped bead image). If you are able to share any images I can give it a try. On Wed, May 24, 2017 at 4:54 AM, Ash <[hidden email]> wrote: > hii > thank you for replying me..yes..i do have multiple bead...the psf has 99 > frames and image has 37 frames....if i am cropping a single bead out then > how should i adjust the size to make it compatible with the raw > image???because raw images size is 2048*2048 and bead size is 512......if i > crop out a single bead, then the bead file will become more smaller!!! > > On Tue, May 23, 2017 at 8:00 PM, bnorthan [via ImageJ] < > [hidden email]> wrote: > > > Hi Ash > > > > If you can share your image and psf I can try and help you. From your > > description I suspect your image contains multiple beads. To use it as a > > PSF you would have to crop out one bead. > > > > Brian > > > > On Mon, May 15, 2017 at 5:45 PM, Ash <[hidden email] > > <http:///user/SendEmail.jtp?type=node&node=5018778&i=0>> wrote: > > > > > my input image size is 2048*2048*37 and psf size is 512*512*99....how > > > should > > > i resize it?do i have to do fft or ifft to get deconvolution > result..the > > > psf > > > file that i have contain several beads....should i take average of all > > the > > > beads in that file to deconvolution.... or just uploading the psf file > > with > > > sveral beads is fine? > > > i am doing with deconvolution lab software...how should i proceed? > > > > > > > > > > > > -- > > > View this message in context: http://imagej.1557.x6.nabble. > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018724.html > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > > > -- > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > ------------------------------ > > If you reply to this email, your message will be added to the discussion > > below: > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > Module-tp5015776p5018778.html > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=unsubscribe_by_code&node=5015776&code=cGhvZWluaXg5M0BnbWFpbC5jb218NT > AxNTc3NnwtNTkxNTYyNDE=> > > . > > NAML > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > web.template.NabbleNamespace-nabble.view.web.template. > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > instant_email%21nabble%3Aemail.naml> > > > > > > > -- > View this message in context: http://imagej.1557.x6.nabble. > com/Deconvolution-Lab-Batch-Module-tp5015776p5018779.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
so before going to deconvolutionLab plugin, first do we have to convolve the image and its PSF?? And then compare results of image convolved with PSF , with the end result of that blurred image with PSF??? Or are we directly starting to deconvolve image that we have taken and its PSF??? On Sat, May 27, 2017 at 5:19 PM, bnorthan [via ImageJ] <[hidden email]> wrote: Hi Ash |
|
Hi Ash
If I understand you correctly, you need to directly deconvolve the image. The microscope has already approximately convolved the true object with the PSF. If you are able to share the image and bead image, I am happy to try and deconvolve them, and let you know the best approach and settings. Brian On Sun, Jun 25, 2017 at 12:09 AM, Ash <[hidden email]> wrote: > so before going to deconvolutionLab plugin, first do we have to convolve > the image and its PSF?? And then compare results of image convolved with > PSF , with the end result of that blurred image with PSF??? > > Or are we directly starting to deconvolve image that we have taken and its > PSF??? > > On Sat, May 27, 2017 at 5:19 PM, bnorthan [via ImageJ] < > [hidden email]> wrote: > > > Hi Ash > > > > If you are using deconvolution lab, it should automatically resize the > PSF > > in x and y. If I recall correctly deconvolution lab does not resize in > z. > > So you should make the PSF x by y by 37 (z same z in image). Make x and > y > > the right size to contain one bead. Make sure the bead is centered. > > > > I also would subtract the background level (at least subtract the min > > value > > of the cropped bead image). > > > > If you are able to share any images I can give it a try. > > > > On Wed, May 24, 2017 at 4:54 AM, Ash <[hidden email] > > <http:///user/SendEmail.jtp?type=node&node=5018792&i=0>> wrote: > > > > > hii > > > thank you for replying me..yes..i do have multiple bead...the psf has > 99 > > > frames and image has 37 frames....if i am cropping a single bead out > > then > > > how should i adjust the size to make it compatible with the raw > > > image???because raw images size is 2048*2048 and bead size is > > 512......if i > > > crop out a single bead, then the bead file will become more smaller!!! > > > > > > On Tue, May 23, 2017 at 8:00 PM, bnorthan [via ImageJ] < > > > [hidden email] <http:///user/SendEmail.jtp? > type=node&node=5018792&i=1>> > > wrote: > > > > > > > Hi Ash > > > > > > > > If you can share your image and psf I can try and help you. From > your > > > > description I suspect your image contains multiple beads. To use it > > as a > > > > PSF you would have to crop out one bead. > > > > > > > > Brian > > > > > > > > On Mon, May 15, 2017 at 5:45 PM, Ash <[hidden email] > > > > <http:///user/SendEmail.jtp?type=node&node=5018778&i=0>> wrote: > > > > > > > > > my input image size is 2048*2048*37 and psf size is > > 512*512*99....how > > > > > should > > > > > i resize it?do i have to do fft or ifft to get deconvolution > > > result..the > > > > > psf > > > > > file that i have contain several beads....should i take average of > > all > > > > the > > > > > beads in that file to deconvolution.... or just uploading the psf > > file > > > > with > > > > > sveral beads is fine? > > > > > i am doing with deconvolution lab software...how should i proceed? > > > > > > > > > > > > > > > > > > > > -- > > > > > View this message in context: http://imagej.1557.x6.nabble. > > > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018724.html > > > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > > > > > > > -- > > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > > > > > > > -- > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > > > > > > ------------------------------ > > > > If you reply to this email, your message will be added to the > > discussion > > > > below: > > > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > > > Module-tp5015776p5018778.html > > > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > macro=unsubscribe_by_code&node=5015776&code= > cGhvZWluaXg5M0BnbWFpbC5jb218NT > > > > > AxNTc3NnwtNTkxNTYyNDE=> > > > > . > > > > NAML > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > > > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > > > web.template.NabbleNamespace-nabble.view.web.template. > > > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > > > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > > > instant_email%21nabble%3Aemail.naml> > > > > > > > > > > > > > > > > > > > -- > > > View this message in context: http://imagej.1557.x6.nabble. > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018779.html > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > > > -- > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > ------------------------------ > > If you reply to this email, your message will be added to the discussion > > below: > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > Module-tp5015776p5018792.html > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=unsubscribe_by_code&node=5015776&code=cGhvZWluaXg5M0BnbWFpbC5jb218NT > AxNTc3NnwtNTkxNTYyNDE=> > > . > > NAML > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > web.template.NabbleNamespace-nabble.view.web.template. > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > instant_email%21nabble%3Aemail.naml> > > > > > > > -- > View this message in context: http://imagej.1557.x6.nabble. > com/Deconvolution-Lab-Batch-Module-tp5015776p5018963.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Thank you, I will do that. .. Well can I send it to your mail id? On Jun 25, 2017 6:16 PM, "bnorthan [via ImageJ]" <[hidden email]> wrote: Hi Ash |
|
In reply to this post by bnorthan
i got results..thanks..one more doubt!! my input image was 8 bits..after deconvolution size is becoming 32 bits...how do i convert output images intensity to that of input image?? i want to compare the images...but since intensities are varying i m not able to compare!! On Sun, Jun 25, 2017 at 11:46 PM, Ash Fairy <[hidden email]> wrote:
|
|
Hi Ash
The dynamic range of a deconvolved image will be larger. So the output probably will not scale to 8 bit. Also make sure you click the option 'normalize PSF'. This will make the PSF is normalized to unit energy, and that will mean you can directly compare input and output values. The deconvolved image should have sharper and higher peaks. This is because blurry "wide" features, should now be narrower and more intense. Brian On Wed, Jun 28, 2017 at 7:06 AM, Ash <[hidden email]> wrote: > i got results..thanks..one more doubt!! > my input image was 8 bits..after deconvolution size is becoming 32 > bits...how do i convert output images intensity to that of input image?? > > i want to compare the images...but since intensities are varying i m not > able to compare!! > > On Sun, Jun 25, 2017 at 11:46 PM, Ash Fairy <[hidden email]> wrote: > > > Thank you, I will do that. .. > > Well can I send it to your mail id? > > > > On Jun 25, 2017 6:16 PM, "bnorthan [via ImageJ]" <ml+s1557n5018964h66@n6. > > nabble.com> wrote: > > > >> Hi Ash > >> > >> If I understand you correctly, you need to directly deconvolve the > image. > >> The microscope has already approximately convolved the true object with > >> the > >> PSF. > >> > >> If you are able to share the image and bead image, I am happy to try and > >> deconvolve them, and let you know the best approach and settings. > >> > >> Brian > >> > >> On Sun, Jun 25, 2017 at 12:09 AM, Ash <[hidden email] > >> <http:///user/SendEmail.jtp?type=node&node=5018964&i=0>> wrote: > >> > >> > so before going to deconvolutionLab plugin, first do we have to > >> convolve > >> > the image and its PSF?? And then compare results of image convolved > >> with > >> > PSF , with the end result of that blurred image with PSF??? > >> > > >> > Or are we directly starting to deconvolve image that we have taken and > >> its > >> > PSF??? > >> > > >> > On Sat, May 27, 2017 at 5:19 PM, bnorthan [via ImageJ] < > >> > [hidden email] <http:///user/SendEmail.jtp? > type=node&node=5018964&i=1>> > >> wrote: > >> > > >> > > Hi Ash > >> > > > >> > > If you are using deconvolution lab, it should automatically resize > >> the > >> > PSF > >> > > in x and y. If I recall correctly deconvolution lab does not resize > >> in > >> > z. > >> > > So you should make the PSF x by y by 37 (z same z in image). Make x > >> and > >> > y > >> > > the right size to contain one bead. Make sure the bead is centered. > >> > > > >> > > I also would subtract the background level (at least subtract the > min > >> > > value > >> > > of the cropped bead image). > >> > > > >> > > If you are able to share any images I can give it a try. > >> > > > >> > > On Wed, May 24, 2017 at 4:54 AM, Ash <[hidden email] > >> > > <http:///user/SendEmail.jtp?type=node&node=5018792&i=0>> wrote: > >> > > > >> > > > hii > >> > > > thank you for replying me..yes..i do have multiple bead...the psf > >> has > >> > 99 > >> > > > frames and image has 37 frames....if i am cropping a single bead > >> out > >> > > then > >> > > > how should i adjust the size to make it compatible with the raw > >> > > > image???because raw images size is 2048*2048 and bead size is > >> > > 512......if i > >> > > > crop out a single bead, then the bead file will become more > >> smaller!!! > >> > > > > >> > > > On Tue, May 23, 2017 at 8:00 PM, bnorthan [via ImageJ] < > >> > > > [hidden email] <http:///user/SendEmail.jtp? > >> > type=node&node=5018792&i=1>> > >> > > wrote: > >> > > > > >> > > > > Hi Ash > >> > > > > > >> > > > > If you can share your image and psf I can try and help you. > From > >> > your > >> > > > > description I suspect your image contains multiple beads. To > use > >> it > >> > > as a > >> > > > > PSF you would have to crop out one bead. > >> > > > > > >> > > > > Brian > >> > > > > > >> > > > > On Mon, May 15, 2017 at 5:45 PM, Ash <[hidden email] > >> > > > > <http:///user/SendEmail.jtp?type=node&node=5018778&i=0>> wrote: > >> > > > > > >> > > > > > my input image size is 2048*2048*37 and psf size is > >> > > 512*512*99....how > >> > > > > > should > >> > > > > > i resize it?do i have to do fft or ifft to get deconvolution > >> > > > result..the > >> > > > > > psf > >> > > > > > file that i have contain several beads....should i take > average > >> of > >> > > all > >> > > > > the > >> > > > > > beads in that file to deconvolution.... or just uploading the > >> psf > >> > > file > >> > > > > with > >> > > > > > sveral beads is fine? > >> > > > > > i am doing with deconvolution lab software...how should i > >> proceed? > >> > > > > > > >> > > > > > > >> > > > > > > >> > > > > > -- > >> > > > > > View this message in context: http://imagej.1557.x6.nabble. > >> > > > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018724.html > >> > > > > > Sent from the ImageJ mailing list archive at Nabble.com. > >> > > > > > > >> > > > > > -- > >> > > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > > > > > >> > > > > > >> > > > > -- > >> > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > > > > >> > > > > > >> > > > > ------------------------------ > >> > > > > If you reply to this email, your message will be added to the > >> > > discussion > >> > > > > below: > >> > > > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > >> > > > > Module-tp5015776p5018778.html > >> > > > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > >> > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > >> > > > macro=unsubscribe_by_code&node=5015776&code= > >> > cGhvZWluaXg5M0BnbWFpbC5jb218NT > >> > > > >> > > > AxNTc3NnwtNTkxNTYyNDE=> > >> > > > > . > >> > > > > NAML > >> > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > >> > > > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > >> > > > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > >> > > > web.template.NabbleNamespace-nabble.view.web.template. > >> > > > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > >> > > > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > >> > > > instant_email%21nabble%3Aemail.naml> > >> > > > > > >> > > > > >> > > > > >> > > > > >> > > > > >> > > > -- > >> > > > View this message in context: http://imagej.1557.x6.nabble. > >> > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018779.html > >> > > > Sent from the ImageJ mailing list archive at Nabble.com. > >> > > > > >> > > > -- > >> > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > > > >> > > > >> > > -- > >> > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > > >> > > > >> > > ------------------------------ > >> > > If you reply to this email, your message will be added to the > >> discussion > >> > > below: > >> > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > >> > > Module-tp5015776p5018792.html > >> > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > >> > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > >> > macro=unsubscribe_by_code&node=5015776&code= > cGhvZWluaXg5M0BnbWFpbC5jb218NT > >> > >> > AxNTc3NnwtNTkxNTYyNDE=> > >> > > . > >> > > NAML > >> > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > >> > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > >> > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > >> > web.template.NabbleNamespace-nabble.view.web.template. > >> > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > >> > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > >> > instant_email%21nabble%3Aemail.naml> > >> > > > >> > > >> > > >> > > >> > > >> > -- > >> > View this message in context: http://imagej.1557.x6.nabble. > >> > com/Deconvolution-Lab-Batch-Module-tp5015776p5018963.html > >> > Sent from the ImageJ mailing list archive at Nabble.com. > >> > > >> > -- > >> > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > >> > >> -- > >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > >> > >> ------------------------------ > >> If you reply to this email, your message will be added to the discussion > >> below: > >> http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Mod > >> ule-tp5015776p5018964.html > >> To unsubscribe from Deconvolution Lab "Batch" Module, click here > >> <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=unsubscribe_by_code&node=5015776&code=cGhvZWluaXg5M0BnbWFpbC5jb218NT > AxNTc3NnwtNTkxNTYyNDE=> > >> . > >> NAML > >> <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > web.template.NabbleNamespace-nabble.view.web.template. > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > instant_email%21nabble%3Aemail.naml> > >> > > > > > > > -- > View this message in context: http://imagej.1557.x6.nabble. > com/Deconvolution-Lab-Batch-Module-tp5015776p5018979.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
yes...it has become more sharper...and i have given normalize psf too!! but the intensity of deconvolved image is totally different...deconvolved images are more dark...and when i am trying to adjust the brightness and intensity of true and deconvolved images, they never match... the true image appears to have more brightness...the intensity is between 0 and 255... for the other it is just 0. so how can i compare it... i wanted to check their snr too.. is there any other parameter that i can check to acess the quality improvement?? On Wed, Jun 28, 2017 at 5:05 PM, bnorthan [via ImageJ] <[hidden email]> wrote: Hi Ash |
|
Hi Ash
Without seeing the images, it is hard for me to understand what is happening. A while back someone had a similar problem, and it turned out it was caused by a scale bar that had been drawn on the image. You can send images to my personal e-mail, or better yet post them somewhere publicly. In general the deconvolved image will appear "dimmer" because of the small bright emitters will have relatively high intensities after deconvolution. A few beads or bead like objects in the images often causes this affect. You can reduce the "max" of the display to help visualize the image. You can also look at line profiles of the objects you are interested in. And take a look at the statistics of the image. The total sum should be similar before and after deconvolution. Brian On Wed, Jun 28, 2017 at 8:00 AM, Ash <[hidden email]> wrote: > yes...it has become more sharper...and i have given normalize psf too!! > > but the intensity of deconvolved image is totally different...deconvolved > images are more dark...and when i am trying to adjust the brightness and > intensity of true and deconvolved images, they never match... > > the true image appears to have more brightness...the intensity is between 0 > and 255... for the other it is just 0. > > > so how can i compare it... > i wanted to check their snr too.. > is there any other parameter that i can check to acess the quality > improvement?? > > On Wed, Jun 28, 2017 at 5:05 PM, bnorthan [via ImageJ] < > [hidden email]> wrote: > > > Hi Ash > > > > The dynamic range of a deconvolved image will be larger. So the output > > probably will not scale to 8 bit. > > > > Also make sure you click the option 'normalize PSF'. This will make the > > PSF is normalized to unit energy, and that will mean you can directly > > compare input and output values. > > > > The deconvolved image should have sharper and higher peaks. This is > > because blurry "wide" features, should now be narrower and more intense. > > > > Brian > > > > On Wed, Jun 28, 2017 at 7:06 AM, Ash <[hidden email] > > <http:///user/SendEmail.jtp?type=node&node=5018980&i=0>> wrote: > > > > > i got results..thanks..one more doubt!! > > > my input image was 8 bits..after deconvolution size is becoming 32 > > > bits...how do i convert output images intensity to that of input > image?? > > > > > > i want to compare the images...but since intensities are varying i m > not > > > able to compare!! > > > > > > On Sun, Jun 25, 2017 at 11:46 PM, Ash Fairy <[hidden email] > > <http:///user/SendEmail.jtp?type=node&node=5018980&i=1>> wrote: > > > > > > > Thank you, I will do that. .. > > > > Well can I send it to your mail id? > > > > > > > > On Jun 25, 2017 6:16 PM, "bnorthan [via ImageJ]" > > <ml+s1557n5018964h66@n6. > > > > nabble.com> wrote: > > > > > > > >> Hi Ash > > > >> > > > >> If I understand you correctly, you need to directly deconvolve the > > > image. > > > >> The microscope has already approximately convolved the true object > > with > > > >> the > > > >> PSF. > > > >> > > > >> If you are able to share the image and bead image, I am happy to try > > and > > > >> deconvolve them, and let you know the best approach and settings. > > > >> > > > >> Brian > > > >> > > > >> On Sun, Jun 25, 2017 at 12:09 AM, Ash <[hidden email] > > > >> <http:///user/SendEmail.jtp?type=node&node=5018964&i=0>> wrote: > > > >> > > > >> > so before going to deconvolutionLab plugin, first do we have to > > > >> convolve > > > >> > the image and its PSF?? And then compare results of image > convolved > > > >> with > > > >> > PSF , with the end result of that blurred image with PSF??? > > > >> > > > > >> > Or are we directly starting to deconvolve image that we have taken > > and > > > >> its > > > >> > PSF??? > > > >> > > > > >> > On Sat, May 27, 2017 at 5:19 PM, bnorthan [via ImageJ] < > > > >> > [hidden email] <http:///user/SendEmail.jtp? > > > type=node&node=5018964&i=1>> > > > >> wrote: > > > >> > > > > >> > > Hi Ash > > > >> > > > > > >> > > If you are using deconvolution lab, it should automatically > > resize > > > >> the > > > >> > PSF > > > >> > > in x and y. If I recall correctly deconvolution lab does not > > resize > > > >> in > > > >> > z. > > > >> > > So you should make the PSF x by y by 37 (z same z in image). > > Make x > > > >> and > > > >> > y > > > >> > > the right size to contain one bead. Make sure the bead is > > centered. > > > >> > > > > > >> > > I also would subtract the background level (at least subtract > the > > > min > > > >> > > value > > > >> > > of the cropped bead image). > > > >> > > > > > >> > > If you are able to share any images I can give it a try. > > > >> > > > > > >> > > On Wed, May 24, 2017 at 4:54 AM, Ash <[hidden email] > > > >> > > <http:///user/SendEmail.jtp?type=node&node=5018792&i=0>> wrote: > > > >> > > > > > >> > > > hii > > > >> > > > thank you for replying me..yes..i do have multiple bead...the > > psf > > > >> has > > > >> > 99 > > > >> > > > frames and image has 37 frames....if i am cropping a single > > bead > > > >> out > > > >> > > then > > > >> > > > how should i adjust the size to make it compatible with the > raw > > > >> > > > image???because raw images size is 2048*2048 and bead size is > > > >> > > 512......if i > > > >> > > > crop out a single bead, then the bead file will become more > > > >> smaller!!! > > > >> > > > > > > >> > > > On Tue, May 23, 2017 at 8:00 PM, bnorthan [via ImageJ] < > > > >> > > > [hidden email] <http:///user/SendEmail.jtp? > > > >> > type=node&node=5018792&i=1>> > > > >> > > wrote: > > > >> > > > > > > >> > > > > Hi Ash > > > >> > > > > > > > >> > > > > If you can share your image and psf I can try and help you. > > > From > > > >> > your > > > >> > > > > description I suspect your image contains multiple beads. > To > > > use > > > >> it > > > >> > > as a > > > >> > > > > PSF you would have to crop out one bead. > > > >> > > > > > > > >> > > > > Brian > > > >> > > > > > > > >> > > > > On Mon, May 15, 2017 at 5:45 PM, Ash <[hidden email] > > > >> > > > > <http:///user/SendEmail.jtp?type=node&node=5018778&i=0>> > > wrote: > > > >> > > > > > > > >> > > > > > my input image size is 2048*2048*37 and psf size is > > > >> > > 512*512*99....how > > > >> > > > > > should > > > >> > > > > > i resize it?do i have to do fft or ifft to get > > deconvolution > > > >> > > > result..the > > > >> > > > > > psf > > > >> > > > > > file that i have contain several beads....should i take > > > average > > > >> of > > > >> > > all > > > >> > > > > the > > > >> > > > > > beads in that file to deconvolution.... or just uploading > > the > > > >> psf > > > >> > > file > > > >> > > > > with > > > >> > > > > > sveral beads is fine? > > > >> > > > > > i am doing with deconvolution lab software...how should i > > > >> proceed? > > > >> > > > > > > > > >> > > > > > > > > >> > > > > > > > > >> > > > > > -- > > > >> > > > > > View this message in context: > http://imagej.1557.x6.nabble. > > > > > >> > > > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018724.html > > > >> > > > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > >> > > > > > > > > >> > > > > > -- > > > >> > > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > > > > > > >> > > > > > > > >> > > > > -- > > > >> > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > > > > > >> > > > > > > > >> > > > > ------------------------------ > > > >> > > > > If you reply to this email, your message will be added to > the > > > >> > > discussion > > > >> > > > > below: > > > >> > > > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > > >> > > > > Module-tp5015776p5018778.html > > > >> > > > > To unsubscribe from Deconvolution Lab "Batch" Module, click > > here > > > >> > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > >> > > > macro=unsubscribe_by_code&node=5015776&code= > > > >> > cGhvZWluaXg5M0BnbWFpbC5jb218NT > > > >> > > > > > >> > > > AxNTc3NnwtNTkxNTYyNDE=> > > > >> > > > > . > > > >> > > > > NAML > > > >> > > > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > >> > > > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > > > >> > > > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > > > >> > > > web.template.NabbleNamespace-nabble.view.web.template. > > > >> > > > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > > > >> > > > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > > > >> > > > instant_email%21nabble%3Aemail.naml> > > > >> > > > > > > > >> > > > > > > >> > > > > > > >> > > > > > > >> > > > > > > >> > > > -- > > > >> > > > View this message in context: http://imagej.1557.x6.nabble. > > > >> > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018779.html > > > >> > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > >> > > > > > > >> > > > -- > > > >> > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > > > > >> > > > > > >> > > -- > > > >> > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > > > >> > > > > > >> > > ------------------------------ > > > >> > > If you reply to this email, your message will be added to the > > > >> discussion > > > >> > > below: > > > >> > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > > >> > > Module-tp5015776p5018792.html > > > >> > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > > > >> > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > >> > macro=unsubscribe_by_code&node=5015776&code= > > > cGhvZWluaXg5M0BnbWFpbC5jb218NT > > > >> > > > >> > AxNTc3NnwtNTkxNTYyNDE=> > > > >> > > . > > > >> > > NAML > > > >> > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > >> > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > > > >> > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > > > >> > web.template.NabbleNamespace-nabble.view.web.template. > > > >> > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > > > >> > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > > > >> > instant_email%21nabble%3Aemail.naml> > > > >> > > > > > >> > > > > >> > > > > >> > > > > >> > > > > >> > -- > > > >> > View this message in context: http://imagej.1557.x6.nabble. > > > >> > com/Deconvolution-Lab-Batch-Module-tp5015776p5018963.html > > > >> > Sent from the ImageJ mailing list archive at Nabble.com. > > > >> > > > > >> > -- > > > >> > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > > >> > > > >> -- > > > >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > >> > > > >> > > > >> ------------------------------ > > > >> If you reply to this email, your message will be added to the > > discussion > > > >> below: > > > >> http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch-Mod > > > >> ule-tp5015776p5018964.html > > > >> To unsubscribe from Deconvolution Lab "Batch" Module, click here > > > >> <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > macro=unsubscribe_by_code&node=5015776&code= > cGhvZWluaXg5M0BnbWFpbC5jb218NT > > > > > AxNTc3NnwtNTkxNTYyNDE=> > > > >> . > > > >> NAML > > > >> <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > > > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > > > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > > > web.template.NabbleNamespace-nabble.view.web.template. > > > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > > > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > > > instant_email%21nabble%3Aemail.naml> > > > >> > > > > > > > > > > > > > > > > > > > -- > > > View this message in context: http://imagej.1557.x6.nabble. > > > com/Deconvolution-Lab-Batch-Module-tp5015776p5018979.html > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > > > -- > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > ------------------------------ > > If you reply to this email, your message will be added to the discussion > > below: > > http://imagej.1557.x6.nabble.com/Deconvolution-Lab-Batch- > > Module-tp5015776p5018980.html > > To unsubscribe from Deconvolution Lab "Batch" Module, click here > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=unsubscribe_by_code&node=5015776&code=cGhvZWluaXg5M0BnbWFpbC5jb218NT > AxNTc3NnwtNTkxNTYyNDE=> > > . > > NAML > > <http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp? > macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml& > base=nabble.naml.namespaces.BasicNamespace-nabble.view. > web.template.NabbleNamespace-nabble.view.web.template. > NodeNamespace&breadcrumbs=notify_subscribers%21nabble% > 3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_ > instant_email%21nabble%3Aemail.naml> > > > > > > > -- > View this message in context: http://imagej.1557.x6.nabble. > com/Deconvolution-Lab-Batch-Module-tp5015776p5018981.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

