Grid Stitching - Can't OK image selection when >50 Images to stitch
|
I'm having an issue stitching large numbers of pictures together with Grid Stitching. Basically the window that opens up for my approval is too large to fit on my screen and there is no scroll bar which would allow me scroll down to the end and process the images. I tried breaking up the images into stacks of 40 or so, but this became tedious.
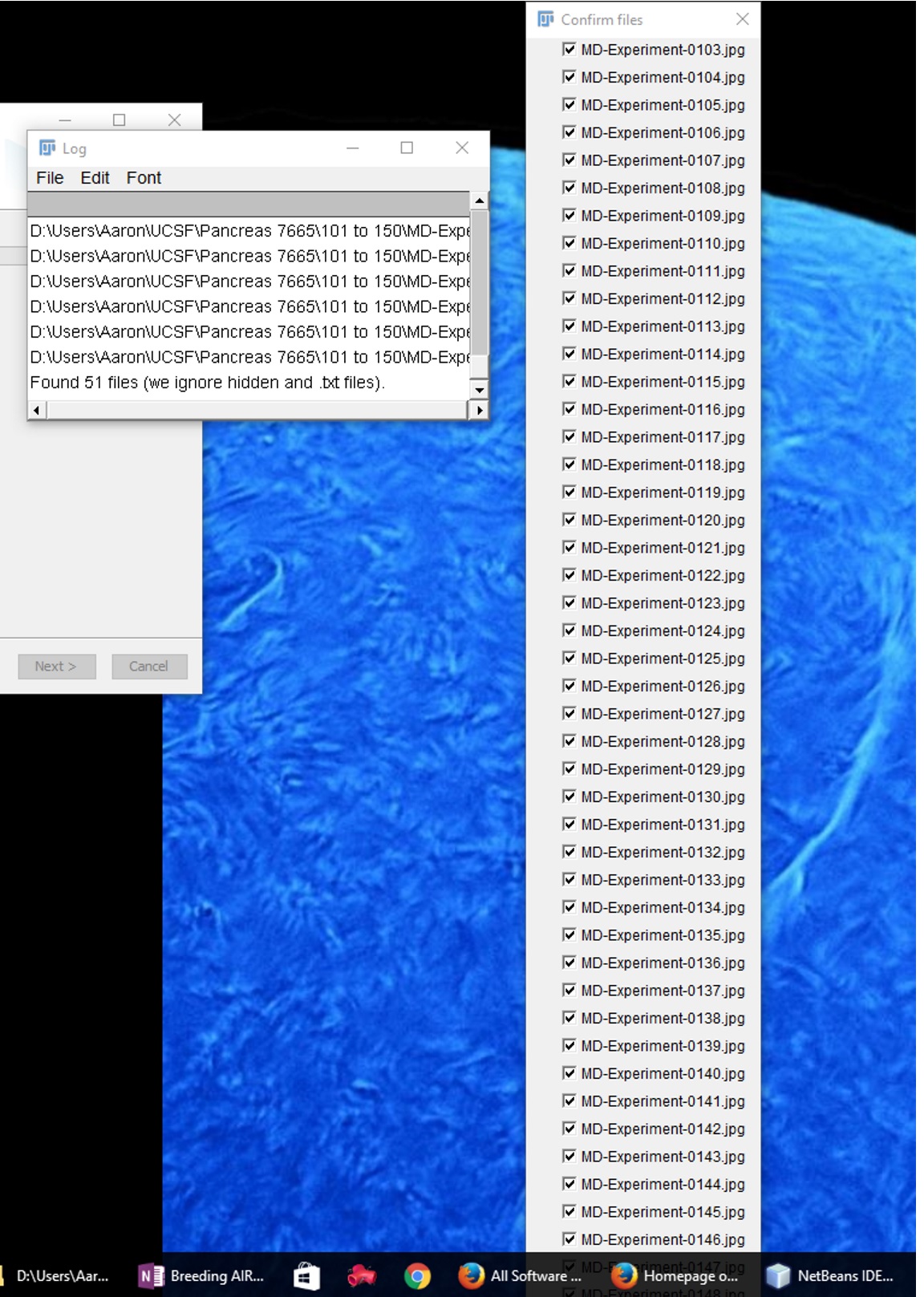 My second attempt to bypass this issue was to write a macro so it would enter in the files for me and I wouldn't have to hit OK. My Macro // Macro for stitching together lots of images. inDir = getDirectory("Choose input directory"); //prompt user to choose input directory outDir = getDirectory("Choose input directory"); // prompt usere to choose save/output directory //Create an array of all files in inDir arr = getFileList(inDir); //Create a string of all file names separated by a space str = ""; for (i=0; i<arr.length; i++) { //Double check that file name is *.tif or *jpg if(endsWith(arr[i], ".jpg")){ str += " " + arr[i]; } if(endsWith(arr[i], ".tif")){ str += " " + arr[i]; } } inDir = replace(inDir, "\\\\$", " "); run("Grid/Collection stitching", "type=[Unknown position] order=[All files in directory] directory=["+inDir+"] confirm_files output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 frame=1 ignore_z_stage display_fusion computation_parameters=[Save computation time (but use more RAM)] image_output=[Fuse and display] "+str+""); But this didn't work. I am not able to process the images this way either. Log File says this: Stitching internal version: 1.2 D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0001.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0002.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0005.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0006.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0010.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0011.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0012.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0013.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0014.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0015.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0016.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0017.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0018.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0019.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0021.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0022.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0023.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0024.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0025.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0026.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0027.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0028.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0029.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0030.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0031.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0032.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0033.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0034.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0035.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0036.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0037.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0038.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0039.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0040.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0041.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0042.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0043.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0044.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0045.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0046.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0047.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0048.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0049.jpg D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0050.jpg Found 44 files (we ignore hidden and .txt files). ERROR: Only 0 files selected, you need at least 2 - stop. ERROR: Error during tile discovery, or invalid grid type. Aborting. Would appreciate some help either with the plugin or with my macro. |
|
Quick solution - run the stitch with sequential images instead of unknown positions. I ran into the same issue when trying to use unknown positions.
If you want to try to get the macro to work, I would record the unknown positions run with only a few images so that you can actually click the confirmation button. Something else may be coming up in the macro that you are not including. Also try only selecting a portion of the images which may reveal a flag in the recorder too. Mike |
|
Thanks Mike,
I wonder what operating system you use? I'm using Windows 10 and for me it doesn't matter if I use sequential or unknown. I don't see a difference in the popup window when choosing either option. I'm still stuck not being able to process all my images. I don't know if a Mac or Linux box would make things different. My screen resolution is 1920x1080 BTW. As to separating them out. I find when stitching together nearly 300 images it becomes very tedious. I then need to break down the images into folders of 40-50 with overlap, run Grid Stitching on each folder, then run the folder with the stitched images (pairwise works better if I don't have enough overlap, but much slower). And then do that again for another 20 data-sets. It's what I have been doing, but I was hoping for a better solution. I recorded a macro for Grid Stitching using only 10 images as the basis of my macro. The recorded macro looked like this: run("Grid/Collection stitching", "type=[Unknown position] order=[All files in directory] directory=[D:\\Users\\Aaron\\UCSF\\Pancreas 7665\\Fused Pics - Copy] confirm_files output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 ignore_z_stage display_fusion computation_parameters=[Save computation time (but use more RAM)] image_output=[Fuse and display] fused_001_to_050.tif fused_051_to_100.tif fused_090_to_110.tif fused_101_to_150.tif fused_140_to_160.tif fused_151_to_200.tif fused_190_to_220.tif fused_201_to_250.tif fused_240_to_260.tif fused_251_to_289.tif"); saveAs("Tiff", "D:\\Users\\Aaron\\UCSF\\Pancreas 7665\\Fused Pics - Copy\\Fused_test.tif"); Then I created variables for both the file path (inDir) and the file names (str) and used them to replace the appropriate areas in the recorded macro. The file path variable (inDir) seems to input correctly, but the image names (str) are not being counted as selected and the Stitching plugin is generating the error in my first post. Thanks for your help again. Aaron |
|
Aaron,
You could also try the grid row by row option, and then just make up a grid that has the right number of tiles. You could even set the overlap to near 100%. There aren't any limitations to how far the stitching program will move tiles, so this might work pretty well. And there is no verification button for that option. Mike |
Re: Grid Stitching - Can't OK image selection when >50 Images to stitch
|
Hi,
an easy way it to try this with a few images where you see the result: - go for all images in directory - once it displays all files, try to navigate with tab (forward) or shift-tab (backward) so the OK button is selected - press Space to run For me the OK button is always selected by default, so just pressing Space works if it does not fit on the screen. Hope this helps, Stephan --- Dr. Stephan Preibisch Group Leader Berlin Institute for Medical Systems Biology (BIMSB), Max Delbrück Center for Molecular Medicine (MDC) Building 89, 1.08b email: [hidden email]<mailto:[hidden email]> web: http://preibischlab.mdc-berlin.de On 14. Apr 2017, at 14:02, MChapman <[hidden email]<mailto:[hidden email]>> wrote: Aaron, You could also try the grid row by row option, and then just make up a grid that has the right number of tiles. You could even set the overlap to near 100%. There aren't any limitations to how far the stitching program will move tiles, so this might work pretty well. And there is no verification button for that option. Mike -- View this message in context: http://imagej.1557.x6.nabble.com/Grid-Stitching-Can-t-OK-image-selection-when-50-Images-to-stitch-tp5018541p5018546.html Sent from the ImageJ mailing list archive at Nabble.com<http://Nabble.com>. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hah! The space bar works great (I was using enter). I still can't scroll through the images and double check them, but it's a great work around.
Also I found removing the "confirm_files" from my macro means I can now macro the process. I'm currently working on making it batch process with automatic saving. I'll post it here when I'm done. Thanks everyone! Aaron. |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

