Help: Save output files to a created directory
|
This post was updated on .
Hello all,
So I am having a little problem with trying to save output files to a created directory and was wondering if anyone might have any ideas as to what the problem may be. My problem is thus, I have created an automated process for collecting tiled images of a slide of Drosophila embryos stained for various markers. The process collects a grid of defined size (6x6) for each embryo and then moves on to the next. The complete output is saved to folder. So, at the end of collection, I am left with a directory of files which is a multiple of 36. With each set of 36 files being one channel for one complete embryo. I am trying to create a macro which will: 1. Choose this directory 2. Run grid/collection stitching plugin for each set of 36 files 3. Save the resulting outputs into distinct folders for each embryo/channel I have gotten pretty far in this process (macro code below), but am running into troubles with File.makeDirectory("path"). In the "for" loop, I have a directory for each channel created and saved as a variable which is called as the output directory required by the Grid/Collection stitching plugin. However, when I go to run the code below, it initiates the plugin, loads the files and then gives me the following error: 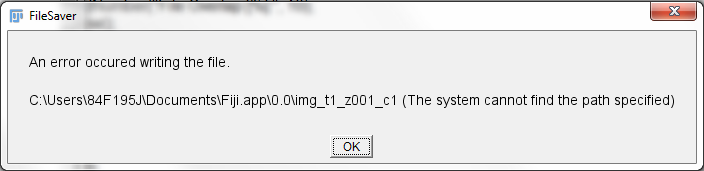 I am completely baffled by why this is occurring. From what I can see, ImageJ is replacing my defined dir488 variable with a nonexistant path "Fiji.app\0.0\". I checked and ImageJ IS creating the directory that I asked it too, so that is not the problem. It just doesn't seem to want to specify it as a path.  In case anyone needs, or wants, to know, the directory with sample images for a test run was placed on my desktop on a Windows 7 PC. Any ideas? I am relatively new to macro programming and have not yet tried anything with directory manipulation, so any advice or help would be greatly appreciated. Sorry in advance for the length and thanks to anyone who reads this through! ===================== Macro code ===================== dir1 = getDirectory("Choose Source Directory "); list = getFileList(dir1); n=list.length; Dialog.create("Parameters"); Dialog.addNumber("Grid Size x:", 6); Dialog.addNumber("Grid Size y:", 6); Dialog.addNumber("File Overlap [%]:", 10); Dialog.show(); xpanels = Dialog.getNumber(); ypanels = Dialog.getNumber(); overlap = Dialog.getNumber(); f = xpanels * ypanels; j = n/f; if ((n%f)!=0) exit("The number of files must be a multiple of "+f+"!"); setBatchMode(true); c=1; for (i=1; i<j; i+=f) { dir488=File.makeDirectory(dir1 + "Embryo_"+c+"\\488\\"); run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & Down ] grid_size_x=&xpanels grid_size_y=&ypanels tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] file_names=[embryo{iii}_w1Confocal 488.tif] output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 computation_parameters=[Save computation time (but use more RAM)] image_output=[Write to disk] output_directory=[&dir488]"); dir647=File.makeDirectory(dir1 + "Embryo_"+c+"\\647\\"); run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & Down ] grid_size_x=&xpanels grid_size_y=&ypanels tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] file_names=[embryo{iii}_w3Confocal 640.tif] output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 computation_parameters=[Save computation time (but use more RAM)] image_output=[Write to disk] output_directory=[&dir647]"); dir405=File.makeDirectory(dir1 + "Embryo_"+c+"\\405\\"); run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & Down ] grid_size_x=&xpanels grid_size_y=&ypanels tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] file_names=[embryo{iii}_w4Confocal 405.tif] output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 computation_parameters=[Save computation time (but use more RAM)] image_output=[Write to disk] output_directory=[&dir405]"); c++; } ===================== Thanks again! *UPDATE* Fixed! Thanks for the useful tips! |
Re: Help: Save output files to a created directory
|
Instead of
dir405=File.makeDirectory(dir1 + "Embryo_"+c+"\\405\\"); try File.makeDirectory(dir1 + "Embryo_"+c+"\\405\\"); dir405 = dir1 + "Embryo_"+c+"\\405\\"; or better: dir405 = dir1 + "Embryo_"+c+"\\405\\"; File.makeDirectory(dir405); File.makeDirectory makes the diretory but does not return a string with the path name. Also, you make need to write: dir405 = dir1+File.separator+"Embryo_"+c+"\\405\\" and perhaps you can replace the \\ with +File.separator+"405" ... (so your code will run on non-windows machines). You can use *File.exists(path)* to check if the directory exists before trying to create it. Here is an example: if (!File.exists(outputPath)) File.makeDirectory(outputPath); hope this helps --aryeh On 1/21/14, 8:31 PM, cnarciso wrote: > Hello all, > > So I am having a little problem with trying to save output files to a > created directory and was wondering if anyone might have any ideas as to > what the problem may be. > > My problem is thus, > > I have created an automated process for collecting tiled images of a slide > of Drosophila embryos stained for various markers. The process collects a > grid of defined size (6x6) for each embryo and then moves on to the next. > The complete output is saved to folder. So, at the end of collection, I am > left with a directory of files which is a multiple of 36. With each set of > 36 files being one channel for one complete embryo. I am trying to create a > macro which will: > > 1. Choose this directory > 2. Run grid/collection stitching plugin for each set of 36 files > 3. Save the resulting outputs into distinct folders for each embryo/channel > > I have gotten pretty far in this process (macro code below), but am running > into troubles with File.makeDirectory("path"). In the "for" loop, I have a > directory for each channel created and saved as a variable which is called > as the output directory required by the Grid/Collection stitching plugin. > However, when I go to run the code below, it initiates the plugin, loads the > files and then gives me the following error: > > <http://imagej.1557.x6.nabble.com/file/n5006211/macroerror.png> > > I am completely baffled by why this is occurring. From what I can see, > ImageJ is replacing my defined dir488 variable with a nonexistant path > "Fiji.app\0.0\". I checked and ImageJ IS creating the directory that I asked > it too, so that is not the problem. It just doesn't seem to want to specify > it as a path. > > In case anyone needs, or wants, to know, the directory with sample images > for a test run was placed on my desktop on a Windows 7 PC. > > Any ideas? > > I am relatively new to macro programming and have not yet tried anything > with directory manipulation, so any advice or help would be greatly > appreciated. Sorry in advance for the length and thanks to anyone who reads > this through! > > ===================== > Macro code > ===================== > dir1 = getDirectory("Choose Source Directory "); > list = getFileList(dir1); > n=list.length; > > Dialog.create("Parameters"); > Dialog.addNumber("Grid Size x:", 6); > Dialog.addNumber("Grid Size y:", 6); > Dialog.addNumber("File Overlap [%]:", 10); > Dialog.show(); > xpanels = Dialog.getNumber(); > ypanels = Dialog.getNumber(); > overlap = Dialog.getNumber(); > > f = xpanels * ypanels; > j = n/f; > if ((n%f)!=0) > exit("The number of files must be a multiple of "+f+"!"); > > setBatchMode(true); > > c=1; > > for (i=1; i<j; i+=f) { > > dir488=File.makeDirectory(dir1 + "Embryo_"+c+"\\488\\"); > > run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & > Down ] grid_size_x=&xpanels grid_size_y=&ypanels > tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] > file_names=[embryo{iii}_w1Confocal 488.tif] > output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] > regression_threshold=0.30 max/avg_displacement_threshold=2.50 > absolute_displacement_threshold=3.50 computation_parameters=[Save > computation time (but use more RAM)] image_output=[Write to disk] > output_directory=[&dir488]"); > > dir647=File.makeDirectory(dir1 + "Embryo_"+c+"\\647\\"); > > run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & > Down ] grid_size_x=&xpanels grid_size_y=&ypanels > tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] > file_names=[embryo{iii}_w3Confocal 640.tif] > output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] > regression_threshold=0.30 max/avg_displacement_threshold=2.50 > absolute_displacement_threshold=3.50 computation_parameters=[Save > computation time (but use more RAM)] image_output=[Write to disk] > output_directory=[&dir647]"); > > dir405=File.makeDirectory(dir1 + "Embryo_"+c+"\\405\\"); > > run("Grid/Collection stitching", "type=[Grid: row-by-row] order=[Right & > Down ] grid_size_x=&xpanels grid_size_y=&ypanels > tile_overlap=&overlap first_file_index_i=&i directory=[&dir1] > file_names=[embryo{iii}_w4Confocal 405.tif] > output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] > regression_threshold=0.30 max/avg_displacement_threshold=2.50 > absolute_displacement_threshold=3.50 computation_parameters=[Save > computation time (but use more RAM)] image_output=[Write to disk] > output_directory=[&dir405]"); > > c++; > > } > ===================== > > Thanks again! > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Help-Save-output-files-to-a-created-directory-tp5006211.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- Aryeh Weiss Faculty of Engineering Bar Ilan University Ramat Gan 52900 Israel Ph: 972-3-5317638 FAX: 972-3-7384051 -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

