Help writing a macro-with pictures!
Help writing a macro-with pictures!
|
Hi everyone,
I don't know much about programming, so I was wondering if someone could help me out. I need to analyze images I have of serotonin receptor clusters (obtained by immunohistochemistry staining and imaged with a confocal-see image at the end of this message). I use the "Find Maxima" function in ImageJ to identify the clusters. The way I want to analyze them is to quantify the cluster size/area and the pixel density of each cluster (to distinguish brighter clusters from dimmer clusters). Since I couldn't find a macro that calculates cluster size and pixel density, I thought it would be easiest to write one. The idea I have for the macro is illustrated in the image below. Only two clusters are shown for clarification purposes. 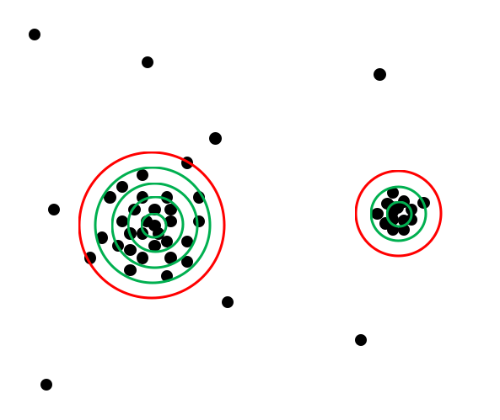 The macro would start by drawing a very small circle around each cluster (determined by "Find Maxima") and calculating the pixel density (number of pixels divided by area of the circle). If the density was above a certain threshold (which would be able to be set by the user), then the program would draw a bigger circle around the same area (green circles in the image). The pixel density of the smaller circle would then be subtracted from the larger, and if this new density is still above threshold, a larger circle is drawn. This process would continue until the pixel density fell below the threshold (red circle in image). In this way, the cluster size can be determined by the area of the biggest circle that was above the pixel density threshold. The pixel density of each cluster can easily be calculated by dividing the number of pixels in the biggest green circle by its area. Here is a typical image that I would use this macro on:  If anyone knows if this program already exists, would be able to help me to write one, or has other suggestions, that would be much appreciated. Thanks for reading my long message! Gabrielle |
|
Hi gabrielle,
if I understand correctly you intend to define the boundary by one measurement of density and give one result also in a measurement of density (albeit a different one). That would worry me a little as I think it would be very very critical what threshold of density was used to define the boundary. Another thing is that pixels are rectangular, not spherical like the dots in your picture. Is each black circle in your picture supposed to be 1 pixel or a large number of pixels? I'm afraid I've not used the find maxima function and haven't read the source code. Is the confocal image at the end of your post before or after using that function? bw eerke |
|
In reply to this post by Gabrielle Van Patten
also how do you propose to designate the centre of each cluster, as opposed to a bright pixel(s) off-centre?
out of interest: what is the hypothesis you are testing? |
Re: Help writing a macro-with pictures!
|
Hey thanks for responding so quickly. I'll try to answer you questions in an efficient way.
We are studying the effects of spinal cord injury on 5HT2A and 5HT2C receptors in mice. Our hypothesis is that because spinal cord injury results in a loss of serotonin (5HT) caudal to the injury site, there is an upregulation in the number and/or size of serotonin receptors.
Each image I have is taken from a specific region of either an intact or spinal cord injured mouse spinal cord, so I want to compare the size, brightness (pixel density) and number of receptors in intact and spinal cord injured mice.
For the density question, yes, I would like to use density to define the boundary and use the total density as part of my results. I think as long is the density threshold is kept constant for the whole image and the images that are being compared, it should be ok. I figured I could just experiment with different thresholds and see what works best.
For my end results, I would like to give the average cluster size, density and total number of clusters per image. Sorry about the pixel confusion. Each dot in my picture was supposed to represent one pixel.
For the find maxima function, if you wanted to try it out yourself in ImageJ, I have attached the .tif version of the image I posted (it's the first one). You just go to process, then binary, then find maxima. I set the noise tolerance by eye, then use the same tolerance for every image I'm comparing the original to.
The only way the image I posted has be altered is I used a deconvolving filter to blur out the background noise. I also attached what the image looks like when you use find maxima (it's the one called point selection).
To designate the center of each cluster, the best way I can think of is to just use the point that the find maxima function designates. That is a good point that the brightest pixels may not be in the center, but I can't think of any other way to do it.
Thank you again for your response. Hopefully I've answered all of your questions and have a nice weekend! Gabrielle
On Sat, Mar 27, 2010 at 3:24 PM, EHB2010 [via ImageJ] <[hidden email]> wrote: also how do you propose to designate the centre of each cluster, as opposed to a bright pixel(s) off-centre? |
|
it won't let me post this as there were too many words containing "a***" so pretend the '*' are 'a'
Hi Gabrielle, I think it might be worth learning a little Java (or possibly macro). It's a lot simpler than you think. To get you started here is a java plug-in that I quickly wrote. I wrote it to be as short, simple and understandable as possible, but at the expense of being slow and not very user friendly. It may also have mistakes; i've only been through it with a debugger once. There are values near the top WRITTEN_LIKE_THIS immediately below "public class SerotoninClusterAn*lyser_ implements PlugIn {". You will want to experiment to see what the best settings are (the number to the right of the '='). apologies if this is too simple. I don't know how much programming you've ever done. THRESHOLD_DENSITY is the really important one. This is the density of positive pixels (black circles on your pic) below which is outside of the cluster. A value of 1 = all pixels in that ring are above threshold brightness. 0 means none. I've set it to 0.3 for no particular reason. RADIUS_INCREMENT is the distance, in pixels, between the green circles in your picture MAXIMUM_NUMBER_OF_RADIUS_INCREMENTS is the maximum number of green circles THRESHOLD_FOR_POSITIVE_FLUORESCENSE. In your picture you have either black circles or white background. this is the threshold that defines a positive (black circle in your pic) pixel; it needs to be a number between 0 (black)and 255 (white). MINIMUM_NUMBER_OF_INCREMENTS. Drawing concentric circles like in your pic is difficult with square pixels if the circle is really small. You might want to start bigger than 1 increment, although I've set the default to 1. I include that setting because it is something to think about and increasing it might need some rewriting of other parts of the code. obviously it is a pain to change these numbers manually and recompile the file each time so you'll probably want to add a graphical interface. There's also a lot of ways of speeding up the plugin and it only uses 1 thread so lots to be worked on. (and there may be mistakes). to run it you need to do your point selection of maxima and then press cmd+m (mac) or ?ctrl+m (pc) to measure them and open the results table. It won't work if you don't do that. You should also make sure the results table is clear before doing the measurement, otherwise you'll confuse the plugin. Then run the plugin but make sure the image you want to an*lyse is the current window or you'll end up analysing the wrong picture. Clearly you could make the whole process much slicker with better code. There's inaccuracy at the margins of the image, because obviously we don't know how bright the pixels outside the image are. You might want to select a region of interest from which "find maxima" that doesn't include the margins of the pic. I've included some download links for the java file and compiled plugin, but the links are not permanent. And don't forget there may be mistakes! ------------start of plugin import ij.plugin.PlugIn; import ij.process.ImageProcessor; import ij.IJ; import ij.ImagePlus; import ij.WindowManager; import ij.measure.*; public class SerotoninClusterAn*lyser_ implements PlugIn { private final static double RADIUS_INCREMENT_IN_PIXELS = 1.5;//you will probably want to change the number private final static int MAXIMUM_NUMBER_OF_RADIUS_INCREMENTS = 25;//you will probably want to change the number private final static byte THRESHOLD_FOR_POSITIVE_FLUORESCENSE = 100;//you will probably want to change the number private final static int MINIMUM_NUMBER_OF_INCREMENTS = 1;//you may want to change the number private final static double THRESHOLD_DENSITY = 0.3; //you may want to change this to value between 0 (none) and 1 (all) private final static String AREA_COLUMN_HEADING = "area (pixels)"; private final static String DENSITY_COLUMN_HEADING = "density (%)"; private final static String MEAN_FLUORESCENCE_OF_CLUSTER = "Mean F"; private ImagePlus imp = WindowManager.getCurrentImage();//get the current image private ResultsTable rt = ResultsTable.getResultsTable();//get the main results table private float[] xCoordinates; private float[] yCoordinates; private int height; private int width; public void run(String arg) { int xColumnNumber = rt.getColumnIndex("X"); int yColumnNumber = rt.getColumnIndex("Y"); if (xColumnNumber != -1 ) {//checks results table is open xCoordinates = rt.getColumn(xColumnNumber); yCoordinates = rt.getColumn(yColumnNumber); } imp = WindowManager.getCurrentImage(); if (imp != null && imp.getBitDepth() == 8 && xCoordinates != null){//only analyse if there is an image and it is 8 bit and there are coordinates doTheProcessing();//start processing } } private final void doTheProcessing() { rt.addColumns();//we need to add two new column for data rt.getFreeColumn(AREA_COLUMN_HEADING); rt.addColumns(); rt.getFreeColumn(DENSITY_COLUMN_HEADING); rt.addColumns(); rt.getFreeColumn(MEAN_FLUORESCENCE_OF_CLUSTER); width = imp.getWidth();//get the image width height = imp.getHeight();//get the image height ImageProcessor byteip = imp.getProcessor();//get the imageprocessor that contains the pixel data double previousRadius = 0; //this bit won't make sense unless you understand nested loops for (int i = 0 ; i < xCoordinates.length ; ++i) { //run through each centre point int aSpecificXCoordinate = (int) xCoordinates[i];//assign the particular x coordinate int aSpecificYCoordinate = (int) yCoordinates[i];//assigns the particular y coordinate double currentOverallDensity = 1;//it starts as 1 as it is the peak pixel int currentOverAllArea = 1;//it is 1 because of the centre pixel being positive; double totalFluorescence = byteip.getPixel(aSpecificXCoordinate, aSpecificYCoordinate); moveToNextCoordinate://break point for when found size for (int j = MINIMUM_NUMBER_OF_INCREMENTS ; j <= MAXIMUM_NUMBER_OF_RADIUS_INCREMENTS; ++j ) { //sequentially adds more radius increments; starts at 1 double currentRadius = j * RADIUS_INCREMENT_IN_PIXELS;//sets current radius int totalPixelsInThisRing = 0; int positivePixelCountInThisRing = 0; double fluorescenceInThisRing = 0; for (int k = 0 ; k < width ; ++k) {//run along the columns in turn for (int l = 0 ; l < height ; ++l) {//run along the rows in turn double distanceCurrentPixelToTheCentrePixel = Math.sqrt(((aSpecificXCoordinate-k)*(aSpecificXCoordinate-k))+((aSpecificYCoordinate-l)*(aSpecificYCoordinate-l))); //now we want to see if the pixel in within the current ring if (distanceCurrentPixelToTheCentrePixel <= currentRadius && distanceCurrentPixelToTheCentrePixel > previousRadius) { totalPixelsInThisRing = totalPixelsInThisRing + 1;//increase pixel count by 1 fluorescenceInThisRing = fluorescenceInThisRing + byteip.getPixel(k,l); if (byteip.getPixel(k,l) > THRESHOLD_FOR_POSITIVE_FLUORESCENSE ){ positivePixelCountInThisRing = positivePixelCountInThisRing + 1;//increment positive pixel count } } } } // now calculate the density in this ring double density = positivePixelCountInThisRing/(totalPixelsInThisRing*1.0); if (density > THRESHOLD_DENSITY) {//if still above threshold include these pixels currentOverallDensity = ((density*totalPixelsInThisRing)+(currentOverallDensity*currentOverAllArea))/(currentOverAllArea+totalPixelsInThisRing); currentOverAllArea = currentOverAllArea + totalPixelsInThisRing; totalFluorescence = totalFluorescence + fluorescenceInThisRing; } else { //else measurements in table and move to next maximum point. int currentCount = rt.getCounter(); if (currentCount < i+1 ) {//just checks we are not going to go out of bounds. probably unnecessary but I don't understand the counter rt.incrementCounter(); } if (currentOverAllArea == 1) { //ie there is only 1 pixel in this cluster currentOverallDensity = 1; //if there is only 1 pixel then our percentage is clearly 100 } rt.setValue(AREA_COLUMN_HEADING, i, (double) currentOverAllArea);//sets the table rt.setValue(DENSITY_COLUMN_HEADING, i, currentOverallDensity*100);//sets the table rt.setValue(MEAN_FLUORESCENCE_OF_CLUSTER, i, totalFluorescence/currentOverAllArea); IJ.showProgress(i/(xCoordinates.length-1.0)); break moveToNextCoordinate; } } } rt.show("Results"); } } --------end plugin downloadable from : http://dl.dropbox.com/u/2014897/SerotoninClusterAnalyser_.class http://dl.dropbox.com/u/2014897/SerotoninClusterAnalyser_.java |
Re: Help writing a macro-with pictures!
|
Wow thank you so much for doing this! I'm sorry I haven't replied sooner. I don't think I got an email saying that anyone replied, so I haven't check back on my post until now. I will definitely try this out and let you know how it goes. Thank you for being so helpful!!
Gabrielle |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

