Linking ROI within ROI to one another (and getting an output)
Linking ROI within ROI to one another (and getting an output)
|
This is a repost (I originally posted a few weeks ago but I don't think the message was ever sent to the mailing list).
I am working on a macro using my limited knowledge of ImageJ in order to analyze cell nuclei and specific features within the nuclei. I am thresholding images twice, the first time to get a general ROI that surrounds a cell's nucleus, and then I am reverting the image and rethresholding within the bounds of the original ROIs in order to get smaller ROIs that surround the nuclear holes (of which there can be multiple within each nucleus). The ultimate goal is to be able to get measurement data for both the nuclei and the holes, which I have had success doing, but I can't seem to figure out an easy way to link the nuclear holes to their "parent" nuclei. Is there any way that FIJI/ImageJ can recognize an ROI within an ROI and provide some sort of output? 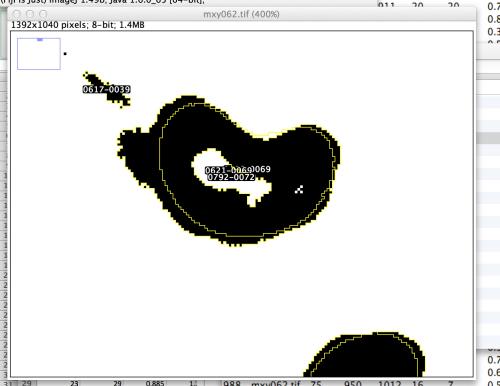 Thanks, - Danny |
|
Hi Danny,
You could try if the Speckle Inspector from the BioVoxxel Toolbox might help you (http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector). This plugin is specifically designed to count small features inside bigger ones. This is first of all only for counting. I will think about including a more detailed analysis for more measurements besides the simple count statistics. regards, Jan 2014-07-10 18:28 GMT+02:00 dprosenberg <[hidden email]>: > This is a repost (I originally posted a few weeks ago but I don't think the > message was ever sent to the mailing list). > > I am working on a macro using my limited knowledge of ImageJ in order to > analyze cell nuclei and specific features within the nuclei. > > I am thresholding images twice, the first time to get a general ROI that > surrounds a cell's nucleus, and then I am reverting the image and > rethresholding within the bounds of the original ROIs in order to get > smaller ROIs that surround the nuclear holes (of which there can be > multiple > within each nucleus). The ultimate goal is to be able to get measurement > data for both the nuclei and the holes, which I have had success doing, but > I can't seem to figure out an easy way to link the nuclear holes to their > "parent" nuclei. > > Is there any way that FIJI/ImageJ can recognize an ROI within an ROI and > provide some sort of output? > > <http://imagej.1557.x6.nabble.com/file/n5008670/Untitled_1.jpg> > > Thanks, > - Danny > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by dprosenberg
Hi Danny,
actually I forgot in my last reply to the mailing list, that you can use the RoiManager output after the Speckle Inspector to run the Analyze Particles on each individual Roi. You might be able to do this with a macro like this: run("Set Measurements...", "area mean standard modal min centroid center perimeter bounding fit shape feret's integrated median skewness kurtosis area_fraction stack display redirect=None decimal=3"); counter = roiManager("count"); for(n=0; n<counter;n++) { roiManager("select", n); run("Analyze Particles...", "display"); } regards, Jan 2014-07-10 18:28 GMT+02:00 dprosenberg <[hidden email]>: > This is a repost (I originally posted a few weeks ago but I don't think the > message was ever sent to the mailing list). > > I am working on a macro using my limited knowledge of ImageJ in order to > analyze cell nuclei and specific features within the nuclei. > > I am thresholding images twice, the first time to get a general ROI that > surrounds a cell's nucleus, and then I am reverting the image and > rethresholding within the bounds of the original ROIs in order to get > smaller ROIs that surround the nuclear holes (of which there can be > multiple > within each nucleus). The ultimate goal is to be able to get measurement > data for both the nuclei and the holes, which I have had success doing, but > I can't seem to figure out an easy way to link the nuclear holes to their > "parent" nuclei. > > Is there any way that FIJI/ImageJ can recognize an ROI within an ROI and > provide some sort of output? > > <http://imagej.1557.x6.nabble.com/file/n5008670/Untitled_1.jpg> > > Thanks, > - Danny > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Danny, hi everyone,
I further updated the "Speckle Inspector <http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector>". Now you can also get a detailed analysis of all small objects in each ROI (from a bigger object) in a results table if desired. I am happy about any feedback. regards, Jan 2014-07-10 21:56 GMT+02:00 BioVoxxel <[hidden email]>: > Hi Danny, > > actually I forgot in my last reply to the mailing list, that you can use > the RoiManager output after the Speckle Inspector to run the Analyze > Particles on each individual Roi. You might be able to do this with a macro > like this: > > run("Set Measurements...", "area mean standard modal min centroid center > perimeter bounding fit shape feret's integrated median skewness kurtosis > area_fraction stack display redirect=None decimal=3"); > counter = roiManager("count"); > for(n=0; n<counter;n++) { > roiManager("select", n); > run("Analyze Particles...", "display"); > } > > > regards, > Jan > > > > > > 2014-07-10 18:28 GMT+02:00 dprosenberg <[hidden email]>: > >> This is a repost (I originally posted a few weeks ago but I don't think >> the >> >> message was ever sent to the mailing list). >> >> I am working on a macro using my limited knowledge of ImageJ in order to >> analyze cell nuclei and specific features within the nuclei. >> >> I am thresholding images twice, the first time to get a general ROI that >> surrounds a cell's nucleus, and then I am reverting the image and >> rethresholding within the bounds of the original ROIs in order to get >> smaller ROIs that surround the nuclear holes (of which there can be >> multiple >> within each nucleus). The ultimate goal is to be able to get measurement >> data for both the nuclei and the holes, which I have had success doing, >> but >> I can't seem to figure out an easy way to link the nuclear holes to their >> "parent" nuclei. >> >> Is there any way that FIJI/ImageJ can recognize an ROI within an ROI and >> provide some sort of output? >> >> <http://imagej.1557.x6.nabble.com/file/n5008670/Untitled_1.jpg> >> >> Thanks, >> - Danny >> >> >> >> -- >> View this message in context: >> http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670.html >> Sent from the ImageJ mailing list archive at Nabble.com. >> >> -- >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html >> > > > > -- > > CEO: Dr. rer. nat. Jan Brocher > phone: +49 (0)6234 917 03 39 > mobile: +49 (0)176 705 746 81 > e-mail: [hidden email] > info: [hidden email] > inquiries: [hidden email] > web: www.biovoxxel.de > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Linking ROI within ROI to one another (and getting an output)
|
Hi Jan, Thank you for getting back to me so quickly! I am working on implementing the plugin and am currently trying to figure out how to use the plugin while running the macro in batch mode (the plugin requires selecting two separate files, both of which must be slightly different variations of one another). Do you have any ideas?
Thank you again!! - Danny Rosenberg - On Fri, Jul 11, 2014 at 6:46 AM, gankaku [via ImageJ] <[hidden email]> wrote:
|
|
Hi Danny
You need the image with your small features you want to analyze per ROI as binary image and another binary image which corresponds to your ROIs. You can get this by either processing your image in a way which (after applying an Autothreshold) will give you the desired binary output or (if this is not possible) you can create your ROIs manually, transfer them to a black image of the same size as the first one and simply fill your ROIs with the white foreground color. Those two images you can then use as input for the Speckle Inspector. Please check out the explanation for it, there you will see, how those two images have to look like (http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector). Alternatively, post an example image and we might be able to help you with further concrete suggestions. This will be way more helpful for you since it deals with your data and the related difficulties. regards, Jan Am 14.07.201417:23 schrieb "dprosenberg" <[hidden email]>: > Hi Jan, > > Thank you for getting back to me so quickly! I am working on implementing > the plugin and am currently trying to figure out how to use the plugin > while running the macro in batch mode (the plugin requires selecting two > separate files, both of which must be slightly different variations of one > another). Do you have any ideas? > > Thank you again!! > > - Danny Rosenberg - > > > On Fri, Jul 11, 2014 at 6:46 AM, gankaku [via ImageJ] < > [hidden email]> wrote: > > > Hi Danny, hi everyone, > > > > I further updated the "Speckle Inspector > > <http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector>". Now you can also > > get > > a detailed analysis of all small objects in each ROI (from a bigger > > object) > > in a results table if desired. > > > > I am happy about any feedback. > > > > regards, > > Jan > > > > > > 2014-07-10 21:56 GMT+02:00 BioVoxxel <[hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=0>>: > > > > > Hi Danny, > > > > > > actually I forgot in my last reply to the mailing list, that you can > use > > > the RoiManager output after the Speckle Inspector to run the Analyze > > > Particles on each individual Roi. You might be able to do this with a > > macro > > > like this: > > > > > > run("Set Measurements...", "area mean standard modal min centroid > center > > > perimeter bounding fit shape feret's integrated median skewness > kurtosis > > > area_fraction stack display redirect=None decimal=3"); > > > counter = roiManager("count"); > > > for(n=0; n<counter;n++) { > > > roiManager("select", n); > > > run("Analyze Particles...", "display"); > > > } > > > > > > > > > regards, > > > Jan > > > > > > > > > > > > > > > > > > 2014-07-10 18:28 GMT+02:00 dprosenberg <[hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=1>>: > > > > > >> This is a repost (I originally posted a few weeks ago but I don't > think > > >> the > > >> > > >> message was ever sent to the mailing list). > > >> > > >> I am working on a macro using my limited knowledge of ImageJ in order > > to > > >> analyze cell nuclei and specific features within the nuclei. > > >> > > >> I am thresholding images twice, the first time to get a general ROI > > that > > >> surrounds a cell's nucleus, and then I am reverting the image and > > >> rethresholding within the bounds of the original ROIs in order to get > > >> smaller ROIs that surround the nuclear holes (of which there can be > > >> multiple > > >> within each nucleus). The ultimate goal is to be able to get > > measurement > > >> data for both the nuclei and the holes, which I have had success > doing, > > >> but > > >> I can't seem to figure out an easy way to link the nuclear holes to > > their > > >> "parent" nuclei. > > >> > > >> Is there any way that FIJI/ImageJ can recognize an ROI within an ROI > > and > > >> provide some sort of output? > > >> > > >> <http://imagej.1557.x6.nabble.com/file/n5008670/Untitled_1.jpg> > > >> > > >> Thanks, > > >> - Danny > > >> > > >> > > >> > > >> -- > > >> View this message in context: > > >> > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670.html > > >> Sent from the ImageJ mailing list archive at Nabble.com. > > >> > > >> -- > > >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > >> > > > > > > > > > > > > -- > > > > > > CEO: Dr. rer. nat. Jan Brocher > > > phone: +49 (0)6234 917 03 39 > > > mobile: +49 (0)176 705 746 81 > > > e-mail: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=2> > > > info: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=3> > > > inquiries: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=4> > > > web: www.biovoxxel.de > > > > > > > > > > > -- > > > > CEO: Dr. rer. nat. Jan Brocher > > phone: +49 (0)6234 917 03 39 > > mobile: +49 (0)176 705 746 81 > > e-mail: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=5> > > info: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=6> > > inquiries: [hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008684&i=7> > > web: www.biovoxxel.de > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > ------------------------------ > > If you reply to this email, your message will be added to the discussion > > below: > > > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008684.html > > To unsubscribe from Linking ROI within ROI to one another (and getting > an > > output), click here > > < > > > > . > > NAML > > < > http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp?macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml&base=nabble.naml.namespaces.BasicNamespace-nabble.view.web.template.NabbleNamespace-nabble.view.web.template.NodeNamespace&breadcrumbs=notify_subscribers%21nabble%3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_instant_email%21nabble%3Aemail.naml > > > > > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008713.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Linking ROI within ROI to one another (and getting an output)
|
Jan, The plugin seems to be working really well, and looks like a much better solution to what I am working on than what I have been currently doing. I think I can figure out how to build a macro that properly creates two binary images the way I need them, and then runs the plugin, but I am not sure how to implement the macro on a wider scale when using batch mode (in other words, each image will have to be duplicated and processed slightly differently before being analyzed via Speckle Inspector), which means the macro will need to know each new file's unique name.
- Danny On Mon, Jul 14, 2014 at 3:25 PM, gankaku [via ImageJ] <[hidden email]> wrote:
|
|
Hi Danny,
To tell a macro to use a specific image for the next step(s) the best is to read its name in a variable as follows: variable = getTitle(); (where variable can be any word you want without spaces) Thereafter you can select it with: selectWindow(variable); Once this is defined, you can always use this variable to select one image or put it as a parameter into a call for a plugin, e.g.: if you have read in the images as variable image1 and image2 you can use the following. run("Speckle Inspector", "big=["+image1+"] small=["+image2+"] min_object=200 min_speckle_number=5 max_speckle_number=22 min_speckle_size=2 exclude individual_roi"); Also see http://rsbweb.nih.gov/ij/developer/macro/functions.html for macro instructions. cheers, Jan 2014-07-14 21:46 GMT+02:00 dprosenberg <[hidden email]>: > Jan, > > The plugin seems to be working really well, and looks like a much better > solution to what I am working on than what I have been currently doing. I > think I can figure out how to build a macro that properly creates two > binary images the way I need them, and then runs the plugin, but I am not > sure how to implement the macro on a wider scale when using batch mode (in > other words, each image will have to be duplicated and processed slightly > differently before being analyzed via Speckle Inspector), which means the > macro will need to know each new file's unique name. > > - Danny > > > > On Mon, Jul 14, 2014 at 3:25 PM, gankaku [via ImageJ] < > [hidden email]> wrote: > > > Hi Danny > > > > You need the image with your small features you want to analyze per ROI > as > > binary image and another binary image which corresponds to your ROIs. You > > can get this by either processing your image in a way which (after > > applying > > an Autothreshold) will give you the desired binary output or (if this is > > not possible) you can create your ROIs manually, transfer them to a black > > image of the same size as the first one and simply fill your ROIs with > the > > white foreground color. > > > > Those two images you can then use as input for the Speckle Inspector. > > > > Please check out the explanation for it, there you will see, how those > two > > images have to look like ( > > http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector). > > Alternatively, post an example image and we might be able to help you > with > > further concrete suggestions. This will be way more helpful for you since > > it deals with your data and the related difficulties. > > > > regards, > > > > Jan > > > > > > > > > > Am 14.07.201417:23 schrieb "dprosenberg" <[hidden email] > > <http://user/SendEmail.jtp?type=node&node=5008724&i=0>>: > > > > > Hi Jan, > > > > > > Thank you for getting back to me so quickly! I am working on > > implementing > > > the plugin and am currently trying to figure out how to use the plugin > > > while running the macro in batch mode (the plugin requires selecting > two > > > separate files, both of which must be slightly different variations of > > one > > > another). Do you have any ideas? > > > > > > Thank you again!! > > > > > > - Danny Rosenberg - > > > > > > > > > On Fri, Jul 11, 2014 at 6:46 AM, gankaku [via ImageJ] < > > > [hidden email] <http://user/SendEmail.jtp?type=node&node=5008724&i=1>> > > wrote: > > > > > > > Hi Danny, hi everyone, > > > > > > > > I further updated the "Speckle Inspector > > > > <http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector>". Now you can > > also > > > > get > > > > a detailed analysis of all small objects in each ROI (from a bigger > > > > object) > > > > in a results table if desired. > > > > > > > > I am happy about any feedback. > > > > > > > > regards, > > > > Jan > > > > > > > > > > > > 2014-07-10 21:56 GMT+02:00 BioVoxxel <[hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=0>>: > > > > > > > > > Hi Danny, > > > > > > > > > > actually I forgot in my last reply to the mailing list, that you > can > > > use > > > > > the RoiManager output after the Speckle Inspector to run the > Analyze > > > > > Particles on each individual Roi. You might be able to do this with > > a > > > > macro > > > > > like this: > > > > > > > > > > run("Set Measurements...", "area mean standard modal min centroid > > > center > > > > > perimeter bounding fit shape feret's integrated median skewness > > > kurtosis > > > > > area_fraction stack display redirect=None decimal=3"); > > > > > counter = roiManager("count"); > > > > > for(n=0; n<counter;n++) { > > > > > roiManager("select", n); > > > > > run("Analyze Particles...", "display"); > > > > > } > > > > > > > > > > > > > > > regards, > > > > > Jan > > > > > > > > > > > > > > > > > > > > > > > > > > > > > > 2014-07-10 18:28 GMT+02:00 dprosenberg <[hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=1>>: > > > > > > > > > >> This is a repost (I originally posted a few weeks ago but I don't > > > think > > > > >> the > > > > >> > > > > >> message was ever sent to the mailing list). > > > > >> > > > > >> I am working on a macro using my limited knowledge of ImageJ in > > order > > > > to > > > > >> analyze cell nuclei and specific features within the nuclei. > > > > >> > > > > >> I am thresholding images twice, the first time to get a general > ROI > > > > that > > > > >> surrounds a cell's nucleus, and then I am reverting the image and > > > > >> rethresholding within the bounds of the original ROIs in order to > > get > > > > >> smaller ROIs that surround the nuclear holes (of which there can > be > > > > >> multiple > > > > >> within each nucleus). The ultimate goal is to be able to get > > > > measurement > > > > >> data for both the nuclei and the holes, which I have had success > > > doing, > > > > >> but > > > > >> I can't seem to figure out an easy way to link the nuclear holes > to > > > > their > > > > >> "parent" nuclei. > > > > >> > > > > >> Is there any way that FIJI/ImageJ can recognize an ROI within an > > ROI > > > > and > > > > >> provide some sort of output? > > > > >> > > > > >> <http://imagej.1557.x6.nabble.com/file/n5008670/Untitled_1.jpg> > > > > >> > > > > >> Thanks, > > > > >> - Danny > > > > >> > > > > >> > > > > >> > > > > >> -- > > > > >> View this message in context: > > > > >> > > > > > > > > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670.html > > > > >> Sent from the ImageJ mailing list archive at Nabble.com. > > > > >> > > > > >> -- > > > > >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > >> > > > > > > > > > > > > > > > > > > > > -- > > > > > > > > > > CEO: Dr. rer. nat. Jan Brocher > > > > > phone: +49 (0)6234 917 03 39 > > > > > mobile: +49 (0)176 705 746 81 > > > > > e-mail: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=2> > > > > > info: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=3> > > > > > inquiries: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=4> > > > > > web: www.biovoxxel.de > > > > > > > > > > > > > > > > > > > > > -- > > > > > > > > CEO: Dr. rer. nat. Jan Brocher > > > > phone: +49 (0)6234 917 03 39 > > > > mobile: +49 (0)176 705 746 81 > > > > e-mail: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=5> > > > > info: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=6> > > > > inquiries: [hidden email] > > > > <http://user/SendEmail.jtp?type=node&node=5008684&i=7> > > > > web: www.biovoxxel.de > > > > > > > > -- > > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > > > > > > ------------------------------ > > > > If you reply to this email, your message will be added to the > > discussion > > > > below: > > > > > > > > > > > > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008684.html > > > > To unsubscribe from Linking ROI within ROI to one another (and > > getting > > > an > > > > output), click here > > > > < > > > > > > > > > > . > > > > NAML > > > > < > > > > > > http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp?macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml&base=nabble.naml.namespaces.BasicNamespace-nabble.view.web.template.NabbleNamespace-nabble.view.web.template.NodeNamespace&breadcrumbs=notify_subscribers%21nabble%3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_instant_email%21nabble%3Aemail.naml > > > > > > > > > > > > > > > > > > > > > > > > > -- > > > View this message in context: > > > > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008713.html > > > > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > > > -- > > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > > > ------------------------------ > > If you reply to this email, your message will be added to the discussion > > below: > > > > > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008724.html > > To unsubscribe from Linking ROI within ROI to one another (and getting > an > > output), click here > > < > > > > . > > NAML > > < > http://imagej.1557.x6.nabble.com/template/NamlServlet.jtp?macro=macro_viewer&id=instant_html%21nabble%3Aemail.naml&base=nabble.naml.namespaces.BasicNamespace-nabble.view.web.template.NabbleNamespace-nabble.view.web.template.NodeNamespace&breadcrumbs=notify_subscribers%21nabble%3Aemail.naml-instant_emails%21nabble%3Aemail.naml-send_instant_email%21nabble%3Aemail.naml > > > > > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Linking-ROI-within-ROI-to-one-another-and-getting-an-output-tp5008670p5008725.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

