Problem with channel bleed-through after splitting channels from RGB image
|
I have been trying to conduct IF analysis with ImageJ and it seems to be giving me a lot of trouble. I always save my images as .tif and they each have a GFP and DAPI channel to them.
When I split them there is no red channel, as expected, but something funky goes on with the GFP channel. It's almost as if ImageJ bleeds over a lot of the DAPI(blue) channel onto the GFP(green) channel after splitting. Here are some images to showcase what I am talking about: 1) Original RGB  2) After Channel Split a) Blue 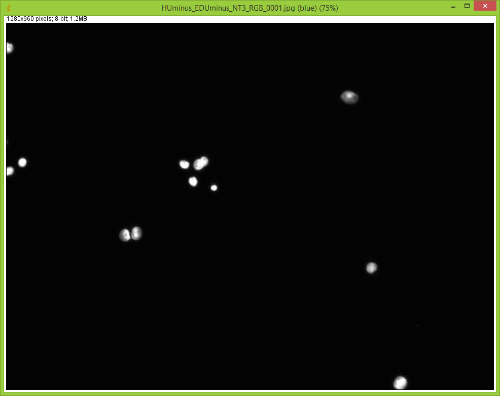 b) Green (I have no clue what is up with the lines, they aren't there when I save the image). 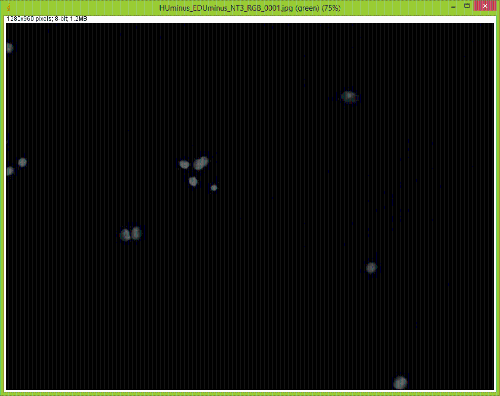 These cells are a negative control and were not treated with EdU. When treated with AlexaFlour (488nm) the EdU present will show up in the GFP channel on the microscope. When imaging these cells there is clearly no signal in the GFP channel but when splitting the channels in ImageJ it creates signal. The only way I have gotten around this is by saving the channels separately but for my negative control it runs into another issue. First, for reference, here are images of non-target cells that have had EdU treatment. As you can imagine there is signal in the GFP channel. 3) Channels saved separately, NOT split through ImageJ a) Blue 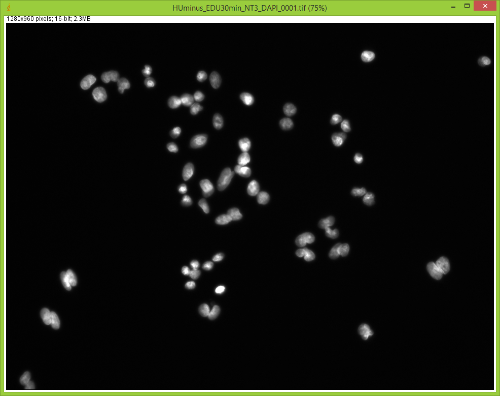 b) Green 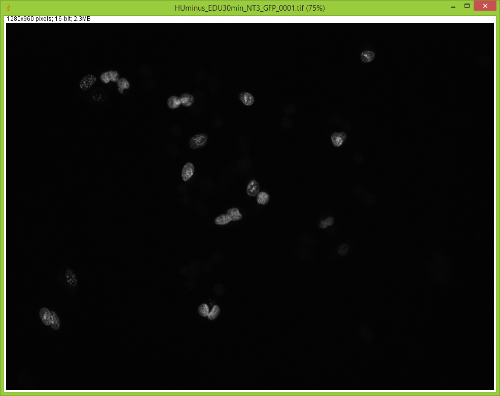 Now here is what happens when I take the RGB image of this and try to split the channels c) Original RGB 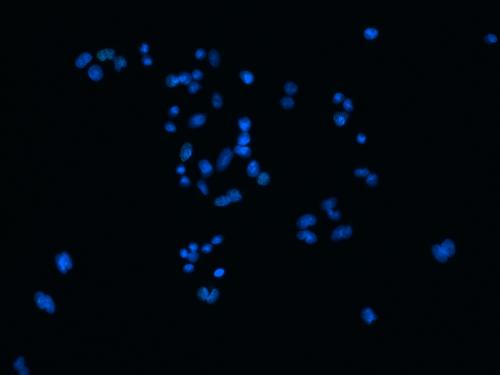 d) Blue 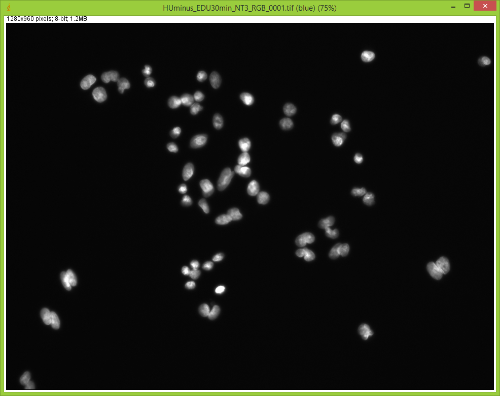 e) Green 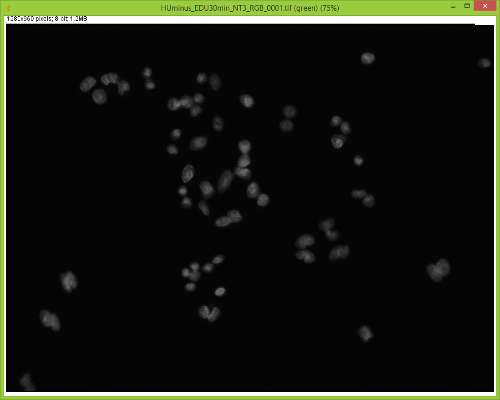 As you can see the green image is simply lower in intensity but ImageJ seems to be unable to separate the DAPI from the image and it completely masks the intensity from the GFP channel. Now here is what happens when I save the cells that have no EdU treatment. 4) Channels saved separately, not split through ImageJ, No EdU a) Blue 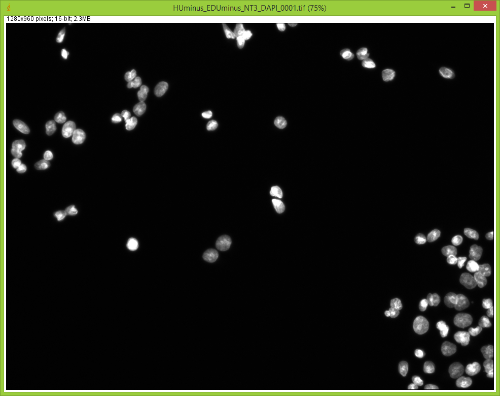 b) Green  I would much rather be able to save all images as just RGB because my macro works perfectly and allows my workflow to be efficient. If I have to switch to the separate channel method I have to re-write the macro and find some way to make it open the GFP image that corresponds to the DAPI one that I choose. I would also need to solve the background issue seen in the negative control (#4). I am willing to learn how to do that but I'd rather find a way that allows me to use what I have now. If anyone has any idea why splitting channels in ImageJ produces bleed-through from other channels that would be great! |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
We have also seen this, and it certainly presents a problem. The DAPI
filter cubes often have a very wide bandpass on the emission side, and so some longer wavelength DAPI signal gets through and is picked up by the camera. One solution is to take images of the three channels separately, separate each of the three images into RGB components, and then reassemble them using the color>merge option. Alternatively, you could look into a narrow bandpass DAPI cube. Joel Joel B. Sheffield, Ph.D Department of Biology Temple University Philadelphia, PA 19122 Voice: 215 204 8839 e-mail: [hidden email] URL: *http://tinyurl.com/khbouft <http://tinyurl.com/khbouft>* On Wed, Nov 4, 2015 at 11:06 AM, FrankyG < [hidden email]> wrote: > I have been trying to conduct IF analysis with ImageJ and it seems to be > giving me a lot of trouble. I always save my images as .tif and they each > have a GFP and DAPI channel to them. > > When I split them there is no red channel, as expected, but something funky > goes on with the GFP channel. It's almost as if ImageJ bleeds over a lot of > the DAPI(blue) channel onto the GFP(green) channel after splitting. > > Here are some images to showcase what I am talking about: > > 1) Original RGB > > < > http://imagej.1557.x6.nabble.com/file/n5014842/HUminus_EDUminus_NT3_RGB_0001.jpg > > > > 2) After Channel Split > a) Blue > > <http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3.png > > > > b) Green (I have no clue what is up with the lines, they aren't there when > I > save the image). > > < > http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3.png > > > > These cells are a negative control and were not treated with EdU. When > treated with AlexaFlour (488nm) the EdU present will show up in the GFP > channel on the microscope. When imaging these cells there is clearly no > signal in the GFP channel but when splitting the channels in ImageJ it > creates signal. > > The only way I have gotten around this is by saving the channels separately > but for my negative control it runs into another issue. > > First, for reference, here are images of non-target cells that have had EdU > treatment. As you can imagine there is signal in the GFP channel. > > 3) Channels saved separately, NOT split through ImageJ > > a) Blue > > < > http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3_Correct.png > > > > b) Green > > < > http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3_Correct.png > > > > Now here is what happens when I take the RGB image of this and try to split > the channels > > c) Original RGB > > < > http://imagej.1557.x6.nabble.com/file/n5014842/HUminus_EDU30min_NT3_RGB_0001_2.jpg > > > > d) Blue > > < > http://imagej.1557.x6.nabble.com/file/n5014842/RGB_EdU30min_Channel_Split_Blue.png > > > > e) Green > > < > http://imagej.1557.x6.nabble.com/file/n5014842/RGB_EdU30min_Channel_Split_Green.png > > > > As you can see the green image is simply lower in intensity but ImageJ > seems > to be unable to separate the DAPI from the image and it completely masks > the > intensity from the GFP channel. > > Now here is what happens when I save the cells that have no EdU treatment. > > 4) Channels saved separately, not split through ImageJ, No EdU > > a) Blue > > < > http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3_Correct_2.png > > > > b) Green > > < > http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3_Correct_2.png > > > > I would much rather be able to save all images as just RGB because my macro > works perfectly and allows my workflow to be efficient. > > If I have to switch to the separate channel method I have to re-write the > macro and find some way to make it open the GFP image that corresponds to > the DAPI one that I choose. I would also need to solve the background issue > seen in the negative control (#4). I am willing to learn how to do that but > I'd rather find a way that allows me to use what I have now. > > If anyone has any idea why splitting channels in ImageJ produces > bleed-through from other channels that would be great! > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Problem-with-channel-bleed-through-after-splitting-channels-from-RGB-image-tp5014842.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
I agree with Joel that DAPI bleeds through like crazy, but I don’t think it’s the problem in this case. A filter issue wouldn't explain why the results are different when the images are saved separately (assuming they were all captured in the same way through the same filters). Instead, I think the problem comes from the default lookup table (LUT) the camera software uses for DAPI. It is not pure blue, but a mix of blue with a little bit of green. I have several software packages that default to some intermediate color like this. Software vendors do this because it makes your “blue” channel easier to see on screen — our eyes are not very sensitive to pure blue. But it causes a problem like you observed when colors are merged in the RGB format. When you split the channels, the “Green” channel now contains your actual green acquisition, plus some of the DAPI signal. For this reason I always recommend to my users to use the Composite format, especially because more of us are using 4+ colors. However, I sympathize with the desire to stick with a successful workflow! So I think you could make it work if you could change the default colorization of the DAPI channel to a pure blue. Hope this helps, Theresa On Nov 4, 2015, at 1:33 PM, JOEL B. SHEFFIELD <[hidden email]<mailto:[hidden email]>> wrote: We have also seen this, and it certainly presents a problem. The DAPI filter cubes often have a very wide bandpass on the emission side, and so some longer wavelength DAPI signal gets through and is picked up by the camera. One solution is to take images of the three channels separately, separate each of the three images into RGB components, and then reassemble them using the color>merge option. Alternatively, you could look into a narrow bandpass DAPI cube. Joel Joel B. Sheffield, Ph.D Department of Biology Temple University Philadelphia, PA 19122 Voice: 215 204 8839 e-mail: [hidden email]<mailto:[hidden email]> URL: *http://tinyurl.com/khbouft <http://tinyurl.com/khbouft>* On Wed, Nov 4, 2015 at 11:06 AM, FrankyG < [hidden email]<mailto:[hidden email]>> wrote: I have been trying to conduct IF analysis with ImageJ and it seems to be giving me a lot of trouble. I always save my images as .tif and they each have a GFP and DAPI channel to them. When I split them there is no red channel, as expected, but something funky goes on with the GFP channel. It's almost as if ImageJ bleeds over a lot of the DAPI(blue) channel onto the GFP(green) channel after splitting. Here are some images to showcase what I am talking about: 1) Original RGB < http://imagej.1557.x6.nabble.com/file/n5014842/HUminus_EDUminus_NT3_RGB_0001.jpg 2) After Channel Split a) Blue <http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3.png b) Green (I have no clue what is up with the lines, they aren't there when I save the image). < http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3.png These cells are a negative control and were not treated with EdU. When treated with AlexaFlour (488nm) the EdU present will show up in the GFP channel on the microscope. When imaging these cells there is clearly no signal in the GFP channel but when splitting the channels in ImageJ it creates signal. The only way I have gotten around this is by saving the channels separately but for my negative control it runs into another issue. First, for reference, here are images of non-target cells that have had EdU treatment. As you can imagine there is signal in the GFP channel. 3) Channels saved separately, NOT split through ImageJ a) Blue < http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3_Correct.png b) Green < http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3_Correct.png Now here is what happens when I take the RGB image of this and try to split the channels c) Original RGB < http://imagej.1557.x6.nabble.com/file/n5014842/HUminus_EDU30min_NT3_RGB_0001_2.jpg d) Blue < http://imagej.1557.x6.nabble.com/file/n5014842/RGB_EdU30min_Channel_Split_Blue.png e) Green < http://imagej.1557.x6.nabble.com/file/n5014842/RGB_EdU30min_Channel_Split_Green.png As you can see the green image is simply lower in intensity but ImageJ seems to be unable to separate the DAPI from the image and it completely masks the intensity from the GFP channel. Now here is what happens when I save the cells that have no EdU treatment. 4) Channels saved separately, not split through ImageJ, No EdU a) Blue < http://imagej.1557.x6.nabble.com/file/n5014842/Blue_Channel_Test_NT-3_Correct_2.png b) Green < http://imagej.1557.x6.nabble.com/file/n5014842/Green_Channel_Test_NT-3_Correct_2.png I would much rather be able to save all images as just RGB because my macro works perfectly and allows my workflow to be efficient. If I have to switch to the separate channel method I have to re-write the macro and find some way to make it open the GFP image that corresponds to the DAPI one that I choose. I would also need to solve the background issue seen in the negative control (#4). I am willing to learn how to do that but I'd rather find a way that allows me to use what I have now. If anyone has any idea why splitting channels in ImageJ produces bleed-through from other channels that would be great! -- View this message in context: http://imagej.1557.x6.nabble.com/Problem-with-channel-bleed-through-after-splitting-channels-from-RGB-image-tp5014842.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html ------------------------------------ Theresa Swayne, Ph.D. Manager Confocal and Specialized Microscopy Shared Resource<http://hiccc.columbia.edu/research/sharedresources/confocal> Herbert Irving Comprehensive Cancer Center Columbia University Medical Center 1130 St. Nicholas Ave., Room 222A New York, NY 10032 Phone: 212-851-4613 [hidden email]<mailto:[hidden email]> -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Joel Sheffield
Dr. Sheffield,
Our scope is a bit different in that for each channel a different light source is used. It still filters the light a bit but the DAPI light source is different than the GFP light source. Unless I understood you wrong and you were not saying that the cubes all filter from one main light source. I will try your method of saving and then merging the files again. Though it requires more macros I'm all for efficient workflow so it will get done somehow. Dr. Swayne, I will check with the EVOS people if it is possible to change the LUT used. If it is possible and that solves the problem than you have many thanks from me for saving me from a lot of extra programming. Thanks for the answers, it's time to get back to work! I will post if I have further issues. |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
On Thursday 05 Nov 2015 07:18:21 FrankyG wrote:
> Dr. Sheffield, > > Our scope is a bit different in that for each channel a different light > source is used. It still filters the light a bit but the DAPI light source > is different than the GFP light source. Unless I understood you wrong and > you were not saying that the cubes all filter from one main light source. > > I will try your method of saving and then merging the files again. Though it > requires more macros I'm all for efficient workflow so it will get done I think there is a much more important problem here. Load your original RGB image of the blue nuclei into IJ and convert it to a RGB stack. Then use the B&C applet to increase the contrast of the channels. You will realise that there is a large number of serious artifacts in your image. Not sure what that is, perhaps you saved the file in jpeg format first (you should never use the jpeg format for any serious imaging). Perhaps you need to recapture the images and make sure that you are saving in a non-lossy format. Regards Gabriel -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
In reply to this post by FrankyG
Hi,
It is not a matter of excitation so much as emission, and the way that images are recorded. Even if you illuminate DAPI with a relatively narrow exciting wavelength, the emission is over a broad range. Look, for instance, at this spectrum (not by me), www.botanik1.uni-karlsruhe.de <http://r.search.yahoo.com/_ylt=A0LEVinodztWAngA1scnnIlQ;_ylu=X3oDMTEzZjZpNzAxBGNvbG8DYmYxBHBvcwMxBHZ0aWQDRkZVSUMwXzEEc2VjA3Nj/RV=2/RE=1446766696/RO=10/RU=http%3a%2f%2fwww.botanik1.uni-karlsruhe.de%2fNick-Lab%2f377.php/RK=0/RS=LN2eASsJgEHyENeKaG9oKRpfZ0w-> . You will see significant emission in the range from 500 almost to 600 nm. This is in the green end of the spectrum, and if the filter cube that you use allows these wavelengths to be transmitted, they will be detected by the green channel of your camera. I am assuming that you are using some form of rgb camera, which separates the light into three channels by using an internal filter mask. Joel B. Sheffield, Ph.D Department of Biology Temple University Philadelphia, PA 19122 Voice: 215 204 8839 e-mail: [hidden email] URL: *http://tinyurl.com/khbouft <http://tinyurl.com/khbouft>* On Thu, Nov 5, 2015 at 10:18 AM, FrankyG < [hidden email]> wrote: > Dr. Sheffield, > > Our scope is a bit different in that for each channel a different light > source is used. It still filters the light a bit but the DAPI light source > is different than the GFP light source. Unless I understood you wrong and > you were not saying that the cubes all filter from one main light source. > > I will try your method of saving and then merging the files again. Though > it > requires more macros I'm all for efficient workflow so it will get done > somehow. > > Dr. Swayne, > > I will check with the EVOS people if it is possible to change the LUT used. > If it is possible and that solves the problem than you have many thanks > from > me for saving me from a lot of extra programming. > > > Thanks for the answers, it's time to get back to work! I will post if I > have > further issues. > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Problem-with-channel-bleed-through-after-splitting-channels-from-RGB-image-tp5014842p5014855.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
In reply to this post by FrankyG
Does your Evos have the monochrome camera or the RGB? The Bayer mask on a single chip RGB camera has a lot of overlap between green and blue, so the green detectors always will detect a little bit of the blue, even if its leaking through an the LED module's emission filter. Bayer mask images are OK for qualitative use, some morphometry is possible after channel subtraction, but useless for intensity based quantitation. The Bayer masks also overlap green and red.
Glen MacDonald Core for Communication Research Virginia Merrill Bloedel Hearing Research Center Cellular Morphology Core Center on Human Development and Disability Box 357923 University of Washington Seattle, WA 98195-7923 USA (206) 616-4156 [hidden email] On Nov 5, 2015, at 7:18 AM, FrankyG <[hidden email]> wrote: > Dr. Sheffield, > > Our scope is a bit different in that for each channel a different light > source is used. It still filters the light a bit but the DAPI light source > is different than the GFP light source. Unless I understood you wrong and > you were not saying that the cubes all filter from one main light source. > > I will try your method of saving and then merging the files again. Though it > requires more macros I'm all for efficient workflow so it will get done > somehow. > > Dr. Swayne, > > I will check with the EVOS people if it is possible to change the LUT used. > If it is possible and that solves the problem than you have many thanks from > me for saving me from a lot of extra programming. > > > Thanks for the answers, it's time to get back to work! I will post if I have > further issues. > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Problem-with-channel-bleed-through-after-splitting-channels-from-RGB-image-tp5014842p5014855.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
This post was updated on .
Dr. Landini,
I save all of them in .tif. I'm not sure what artifacts you are referring to and I stopped using the RGB for quantitative analysis because of the issues I realized once I split the images. Saving them as separate channels fixes almost all the issues I have been having. Perhaps look at the images that are separate channels and see if you can find artifacts? Those are the ones I am using now, not the RGB. To be clear they are saved separately from the RGB, perhaps that was not clear. Dr. MacDonald, Yes our EVOS has a monochrome camera and the representative explained it to me. So are you saying that even if I save the channels separately that the imaging system I am using is absolutely useless for quantitative studies? If so I'll have to notify my PI ASAP because I do not think he was aware of that. |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
Dear FrankyG,
You will need to get the specifications for your particular Evos. They come with different configurations and different cameras. You should also have the filter specs available. Your comments suggest 2 cameras on your Evos? If you have a trully monochrome camera, then it should be used for quantitative imaging, collecting each channel sequentially. If you have a CCD with mosaic filters (Bayer mask) there is a modest loss of spatial resolution, overlap between the camera filters means any bleedthrough in the imaging system will be picked up by the other channels, you are limited to 8-bits/pixel and unless you have access to the RAW format, the “colors’ are established by firmware interpolation of intensity neighborhoods. An RGB image from a Bayer mask color camera can be set to gray, but its just a LUT manipulation and you still have the above issues. A monochrome camerea does not need to save as RAW since there is no need to alter the captured intensities, provided you do save it in full bit depth rather than as RGB. However, I haven’t used those scopes in a long time and don’t know what they are up these days with their software. Regards Glen MacDonald Core for Communication Research Virginia Merrill Bloedel Hearing Research Center Cellular Morphology Core Center on Human Development and Disability Box 357923 University of Washington Seattle, WA 98195-7923 USA (206) 616-4156 [hidden email] On Nov 5, 2015, at 1:34 PM, FrankyG <[hidden email]> wrote: > Dr. Landini, > > I save all of them in .tif. > > I'm not sure what artifacts you are referring to and I stopped using the RGB > for quantitative analysis because of the issues I realized once I split the > images. Saving them as separate channels fixes almost all the issues I have > been having. Perhaps look at the images that are separate channels and see > if you can find artifacts? Those are the ones I am using now, not the RGB. > > To be clear they are saved separately from the RGB, perhaps that was not > clear. > > Dr. MacDonald, > > Yes our EVOS has a monochrome camera and the representative explained it to > me. I'm willing to bet it is an RGB camera as well as I cannot save as RAW, > only RGB format. > > So are you saying that even if I save the channels separately that the > imaging system I am using is absolutely useless for quantitative studies? If > so I'll have to notify my PI ASAP because I do not think he was aware of > that. > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Problem-with-channel-bleed-through-after-splitting-channels-from-RGB-image-tp5014842p5014867.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Problem with channel bleed-through after splitting channels from RGB image
|
In reply to this post by FrankyG
On Thursday 05 Nov 2015 13:34:50 FrankyG wrote:
> I save all of them in .tif. Your RGB image might have been saved in tiff format but only after being saved into something else (by you or by the software). For sure it has compression artifacts. Please follow the steps I mentioned in you previous email with the first image you posted and you will see what I mean. Even if you do not intend to use the RGB version, it might be useful to know what is going on to avoid analysing artifacts rather than data. Cheers Gabriel -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
This post was updated on .
I do not have access to the RAW format unfortunately and though I can make it so it doesn't save it as 8-bit, there is no way to save it other than RGB with sequential channels also saved separately.
I'm willing to bet the compression artifacts are from the software and there's nothing I can do about it. Essentially the rep told me that what's there to manipulate is in the options menu on the scope. All I can do from there is save channels separately and ask it to not save everything as 8-bit. I guess this means I will be needing a different scope to do quantitative analysis. |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

