Separating cells close to each other
Separating cells close to each other
|
CONTENTS DELETED
The author has deleted this message.
|
Re: Separating cells close to each other
|
Hi Tom,
one possibility: (1) subtract a background, e.g. using the Fit Polynomial plugin (2) threshold to get all cell outlines red (3) analyze particles with suitable minimum size to avoid noise pixels, Output =mask and 'Include holes' checked (4) Watershed to cut between adjacent cells. It all depends on whether you can get the full cell outlines with thresholding. An alternative method of getting the outlines might be the Versatile Wand with a large value tolerance and gradient limitation (e.g. at roughly 4 for your jpg image). Then you can convert the selection to a mask and run Watershed on it. Fit Polynomial: http://imagejdocu.tudor.lu/doku.php? id=plugin:filter:fit_polynomial:start Versatile Wand: http://imagejdocu.tudor.lu/doku.php? id=plugin:segmentation:versatile_wand:start Michael ________________________________________________________________ On 7 Oct 2011, at 15:21, Tom Runia wrote: > Hi everybody, > > I am working on an automated cell counter (plugin), my images > contain cells > close to each other. I was wondering whether there is (an easy) way to > separate cells close to each other using ImageJ? > Also see this image: > http://tweakers.net/ext/f/TNIar0XBouHtIMnxr0VPkpph/full.jpg > > These are yeast cells with buds, the goal is to count the mother > cell and > bud separately (any ideas and thoughts about this are more than > welcome!). > > gr, > > Tom |
Re: Separating cells close to each other
|
CONTENTS DELETED
The author has deleted this message.
|
Re: Separating cells close to each other
|
In reply to this post by Michael Schmid
Hi Tom and Michael,
I'm trying to do something similar, except with axons in a nerve cross section, with lots of background. I have been using the default "Subtract Background" function within ImageJ, would the Fit Polynomial work better? I'm very new to working with ImageJ. here's a sample image: http://www.swapdrive.com/file.asp?ID=tG+4c+dQUV2uaBUzGE9QXIqvYD6NpPr0zU3HMp4maEzHuo55/04pqQ==url I'm trying to measure the average area of each of the axons within the nerve, including the myelin sheath. What I did so far is: Process>Subtract Background>5 pixels Process>Binary>Make Binary Analyze>Analyze Particles>Size 4-350, circularity 0.25-1.0, show outlines, include holes I chose 350 as max size because that allows me to eliminate blood vessels within the nerve. I arbitrarily chose 0.25 circularity because I didn't know what else to pick. Doing this gave me an average size of 50.668 pixels. I will put a slide with a length marker under microscope with same magnification so I can use "Analyze > Set Scale" and get values in micrometers. Do you guys think this would be an OK way of measuring the axon cross section area? Thank you again! |
Different background subtraction methods (Was: Separating cells close to each other)
|
Hi Ziyi,
it depends on the image type whether 'Fit Polynomial' or 'Subtract Background' are better. Unfortunately, I can't see your image ('maximum downloads exceeded' error). Subtract Background requires that the object are EITHER brighter OR darker than the background. For dark objects, there should be nothing in the image brighter than what you consider background, and for bright objects, nothing in the image should be darker than the background. Fit Polynomial (also the Highpass filter, which subtracts the Gaussian-blurred image) is based on the average brightness. So, if you have large or many objects that are on average significantly brighter or darker than the background, they won't work well. Fit Polynomial takes the background from the selection only. So, if you can get an approximate selection or mask with your objects as a first step (Thresholding, Versatile Wand with gradient detection, or whatever), this will help you: Create the inverse selection (Edit>Selection>Make Inverse) and run Fit Polynomial with that selection (the background). [P.S: Have I guessed your name correctly from the email address? Maybe I am an old-fashioned European who prefers to talk to people with names...] Michael ________________________________________________________________ On 8 Oct 2011, at 03:56, GloryField wrote: > Hi Tom and Michael, > > I'm trying to do something similar, except with axons in a nerve cross > section, with lots of background. > I have been using the default "Subtract Background" function within > ImageJ, > would the Fit Polynomial work better? I'm very new to working with > ImageJ. > > here's a sample image: > http://www.swapdrive.com/file.asp?ID=tG+4c > +dQUV2uaBUzGE9QXIqvYD6NpPr0zU3HMp4maEzHuo55/04pqQ==url > > I'm trying to measure the average area of each of the axons within the > nerve, including the myelin sheath. What I did so far is: > > Process>Subtract Background>5 pixels > Process>Binary>Make Binary > Analyze>Analyze Particles>Size 4-350, circularity 0.25-1.0, show > outlines, > include holes > > I chose 350 as max size because that allows me to eliminate blood > vessels > within the nerve. > I arbitrarily chose 0.25 circularity because I didn't know what > else to > pick. > > Doing this gave me an average size of 50.668 pixels. I will put a > slide > with a length marker under microscope with same magnification so I > can use > "Analyze > Set Scale" and get values in micrometers. > > Do you guys think this would be an OK way of measuring the axon cross > section area? > > Thank you again! > > -- > View this message in context: http://imagej.588099.n2.nabble.com/ > Separating-cells-close-to-each-other-tp6869352p6871669.html > Sent from the ImageJ mailing list archive at Nabble.com. |
Re: Separating cells close to each other
|
In reply to this post by verified.human
Dear Tom & Micheal
I am working on something similar but I am trying to count droplets and determine their size. Here is an image: http://imageshack.us/photo/my-images/52/1p00cimg0067.jpg/ Could I use the same technique? Any help would be much appreciated. Regards Miné On Fri, Oct 7, 2011 at 3:21 PM, Tom Runia <[hidden email]> wrote: > Hi everybody, > > I am working on an automated cell counter (plugin), my images contain cells > close to each other. I was wondering whether there is (an easy) way to > separate cells close to each other using ImageJ? > Also see this image: > http://tweakers.net/ext/f/TNIar0XBouHtIMnxr0VPkpph/full.jpg > > These are yeast cells with buds, the goal is to count the mother cell and > bud separately (any ideas and thoughts about this are more than welcome!). > > gr, > > Tom > |
Re: Separating cells close to each other
|
Hi Miné
if your images have somewhat better resolution than the uploaded one, you might try template matching with the Feature Finder, http://imagejdocu.tudor.lu/doku.php? id=plugin:analysis:feature_finder:start Michael ________________________________________________________________ On 10 Oct 2011, at 15:23, Miné Zantow wrote: > Dear Tom & Micheal > > I am working on something similar but I am trying to count droplets > and > determine their size. > > Here is an image: > > http://imageshack.us/photo/my-images/52/1p00cimg0067.jpg/ > > Could I use the same technique? Any help would be much appreciated. > > Regards > > Miné > > On Fri, Oct 7, 2011 at 3:21 PM, Tom Runia <[hidden email]> wrote: > >> Hi everybody, >> >> I am working on an automated cell counter (plugin), my images >> contain cells >> close to each other. I was wondering whether there is (an easy) >> way to >> separate cells close to each other using ImageJ? >> Also see this image: >> http://tweakers.net/ext/f/TNIar0XBouHtIMnxr0VPkpph/full.jpg >> >> These are yeast cells with buds, the goal is to count the mother >> cell and >> bud separately (any ideas and thoughts about this are more than >> welcome!). >> >> gr, >> >> Tom >> |
Re: Separating cells close to each other
|
Dear Michael
Thank you for your help Miné On Mon, Oct 10, 2011 at 3:52 PM, Michael Schmid <[hidden email]>wrote: > Hi Miné > > if your images have somewhat better resolution than the uploaded one, you > might try template matching with the Feature Finder, > > http://imagejdocu.tudor.lu/**doku.php?id=plugin:analysis:** > feature_finder:start<http://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:feature_finder:start> > > > Michael > ______________________________**______________________________**____ > > > On 10 Oct 2011, at 15:23, Miné Zantow wrote: > > Dear Tom & Micheal >> >> I am working on something similar but I am trying to count droplets and >> determine their size. >> >> Here is an image: >> >> http://imageshack.us/photo/my-**images/52/1p00cimg0067.jpg/<http://imageshack.us/photo/my-images/52/1p00cimg0067.jpg/> >> >> Could I use the same technique? Any help would be much appreciated. >> >> Regards >> >> Miné >> >> On Fri, Oct 7, 2011 at 3:21 PM, Tom Runia <[hidden email]> wrote: >> >> Hi everybody, >>> >>> I am working on an automated cell counter (plugin), my images contain >>> cells >>> close to each other. I was wondering whether there is (an easy) way to >>> separate cells close to each other using ImageJ? >>> Also see this image: >>> http://tweakers.net/ext/f/**TNIar0XBouHtIMnxr0VPkpph/full.**jpg<http://tweakers.net/ext/f/TNIar0XBouHtIMnxr0VPkpph/full.jpg> >>> >>> These are yeast cells with buds, the goal is to count the mother cell and >>> bud separately (any ideas and thoughts about this are more than >>> welcome!). >>> >>> gr, >>> >>> Tom >>> >>> |
Re: Different background subtraction methods (Was: Separating cells close to each other)
|
In reply to this post by Michael Schmid
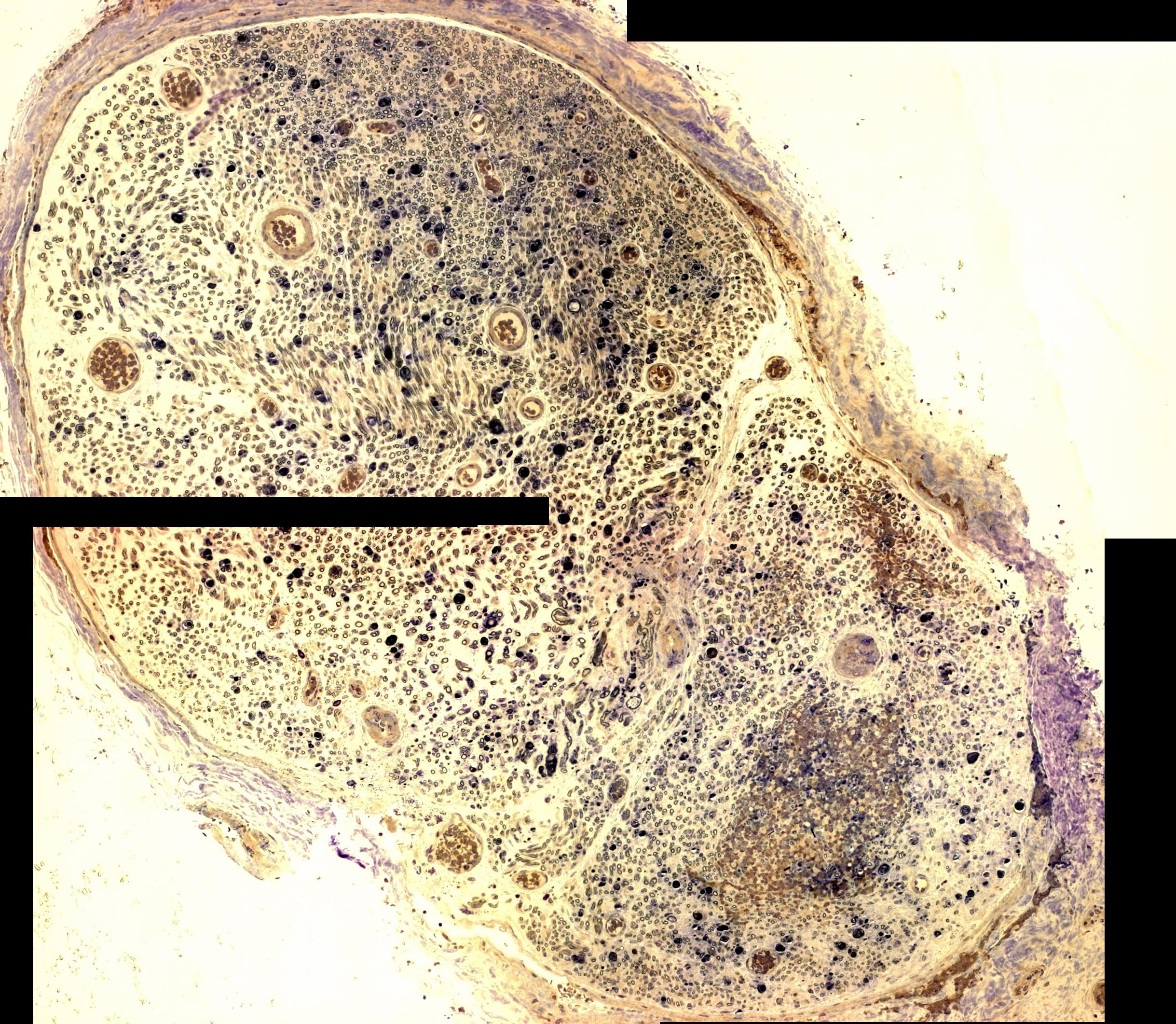 Hi Michael, Yes, my name is Ziyi, good guess! I attached the image to the post this time, hopefully it works. Basically I'm trying to reduce the background so I can do analyze particles and count the area of each of the axons within the nerve. So what I plan on doing is: Reduce Background (or fit polynomial) Make Binary Analyze Particles -> size 5-350, circularity 0.25-1 Do you think this would be OK to give accurate results? I figured since there are over 10,000 axons, the occasional mis-inclusion/exclusion would not affect the results very much. Z |
Re: Different background subtraction methods (Was: Separating cells close to each other)
|
and here are two more sample images:
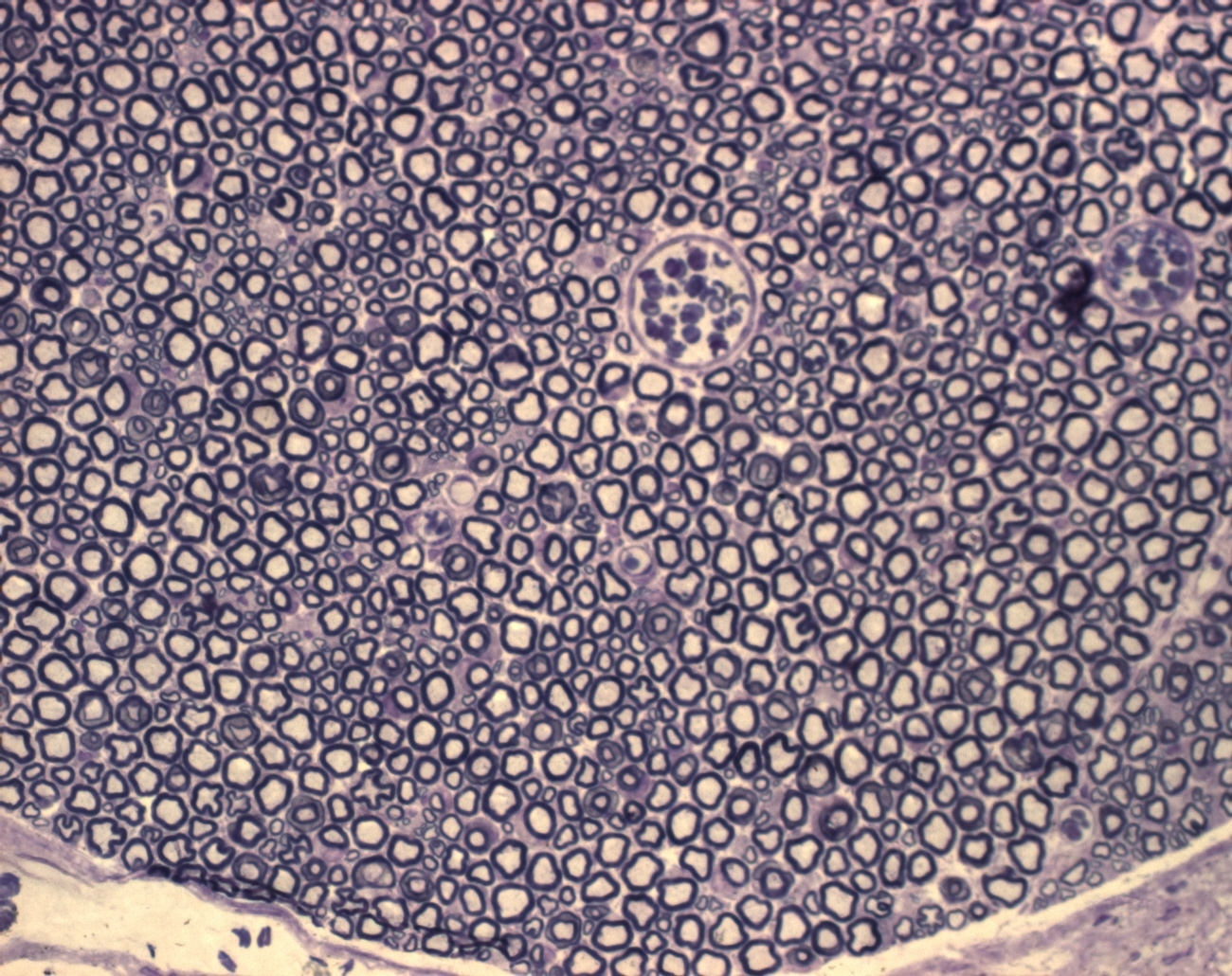 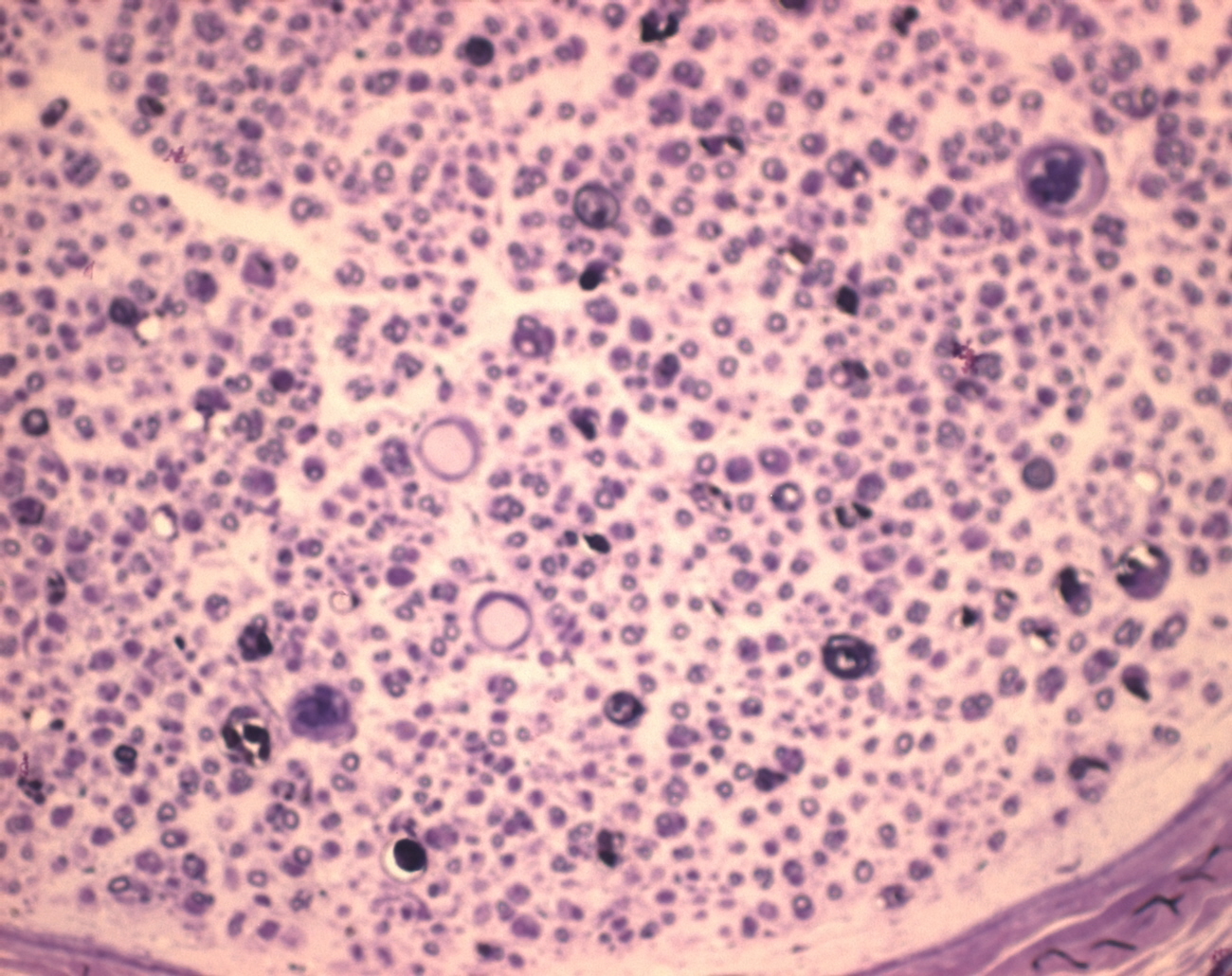
|
Re: Different background subtraction methods (Was: Separating cells close to each other)
|
Years ago we wrote a macro in NIH-Image which did a pretty good job of isolating the axon cross sections and reporting the areas of myelin vs axon. It was mostly a simple thresholding method. It would have worked with B1-CD2 but not with the fuzzy A1-D2. Also, the user would have to manually cut out the blood vessel and sometimes manually draw white lines to separate myelin rings from abutting axons.
-Michael C. -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of GloryField Sent: Monday, October 10, 2011 2:29 PM To: [hidden email] Subject: Re: Different background subtraction methods (Was: Separating cells close to each other) and here are two more sample images: http://imagej.588099.n2.nabble.com/file/n6877934/B1-CD2.jpg http://imagej.588099.n2.nabble.com/file/n6877934/A1-D2.jpg -- View this message in context: http://imagej.588099.n2.nabble.com/Separating-cells-close-to-each-other-tp6869352p6877934.html Sent from the ImageJ mailing list archive at Nabble.com. ------------------------------------------------------------ This email message, including any attachments, is for the sole use of the intended recipient(s) and may contain information that is proprietary, confidential, and exempt from disclosure under applicable law. Any unauthorized review, use, disclosure, or distribution is prohibited. If you have received this email in error please notify the sender by return email and delete the original message. Please note, the recipient should check this email and any attachments for the presence of viruses. The organization accepts no liability for any damage caused by any virus transmitted by this email. ================================= |
Re: Different background subtraction methods (Was: Separating cells close to each other)
|
Hi Michael,
Is there a way to do this in ImageJ, or do you think the Subtract Background/Analyze Particles would be sufficient? We are trying to measure the entire area including the myelin, and comparing the average axon (including myelin) area between different treatment groups. Thanks, Z |
Re: Different background subtraction methods (Was: Separating cells close to each other)
|
In reply to this post by GloryField
Hi Ziyi,
some ideas: D1-D_Composite-1: ================= You have small objects with a bright interior. "Subtract Background' would take that interior level as a background. What you can try: Duplicate the image run("Minimum...", "radius=2"); //removes bright interior run("Subtract Background...", "rolling=20 light separate create sliding"); and subtract this background from the original B1-CD2: ======= You have so many objects of different size and contrast, often touching or intersecting, that it will be difficult to get reliable results. For the background subtraction, maybe try something like this: median r=2 minimum r=3 maximum r=15 mean r=15 minimum r=12 and subtract the result from the original. If you are interested in the ring-shaped objects, you could try threshold with dark background, so you get the inner bright areas, and use suitable filters of the particle analyzer (size, circularity) to eliminate most of the background. A1-D2: ====== This is one where Subtract background works well (e.g. sliding paraboloid with radius=20). Thresholding and separating the objects is difficult, however, because there are many objects of different size, shapes brightness, etc. As I am not in biology or life sciences, I have no idea which of the objects are of interest for you, so I leave this to people who know something about it ;-). Michael ________________________________________________________________ On 10 Oct 2011, at 20:24, GloryField wrote: > http://imagej.588099.n2.nabble.com/file/n6877921/D1-D_Composite.jpg > > Hi Michael, > > Yes, my name is Ziyi, good guess! > > I attached the image to the post this time, hopefully it works. > > Basically I'm trying to reduce the background so I can do analyze > particles > and count the area of each of the axons within the nerve. > > So what I plan on doing is: > Reduce Background (or fit polynomial) > Make Binary > Analyze Particles -> size 5-350, circularity 0.25-1 > > Do you think this would be OK to give accurate results? I figured > since > there are over 10,000 axons, the occasional mis-inclusion/exclusion > would > not affect the results very much. > > Z > > and here are two more sample images: > http://imagej.588099.n2.nabble.com/file/n6877934/B1-CD2.jpg > http://imagej.588099.n2.nabble.com/file/n6877934/A1-D2.jpg |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

