count nuclei in low mag image
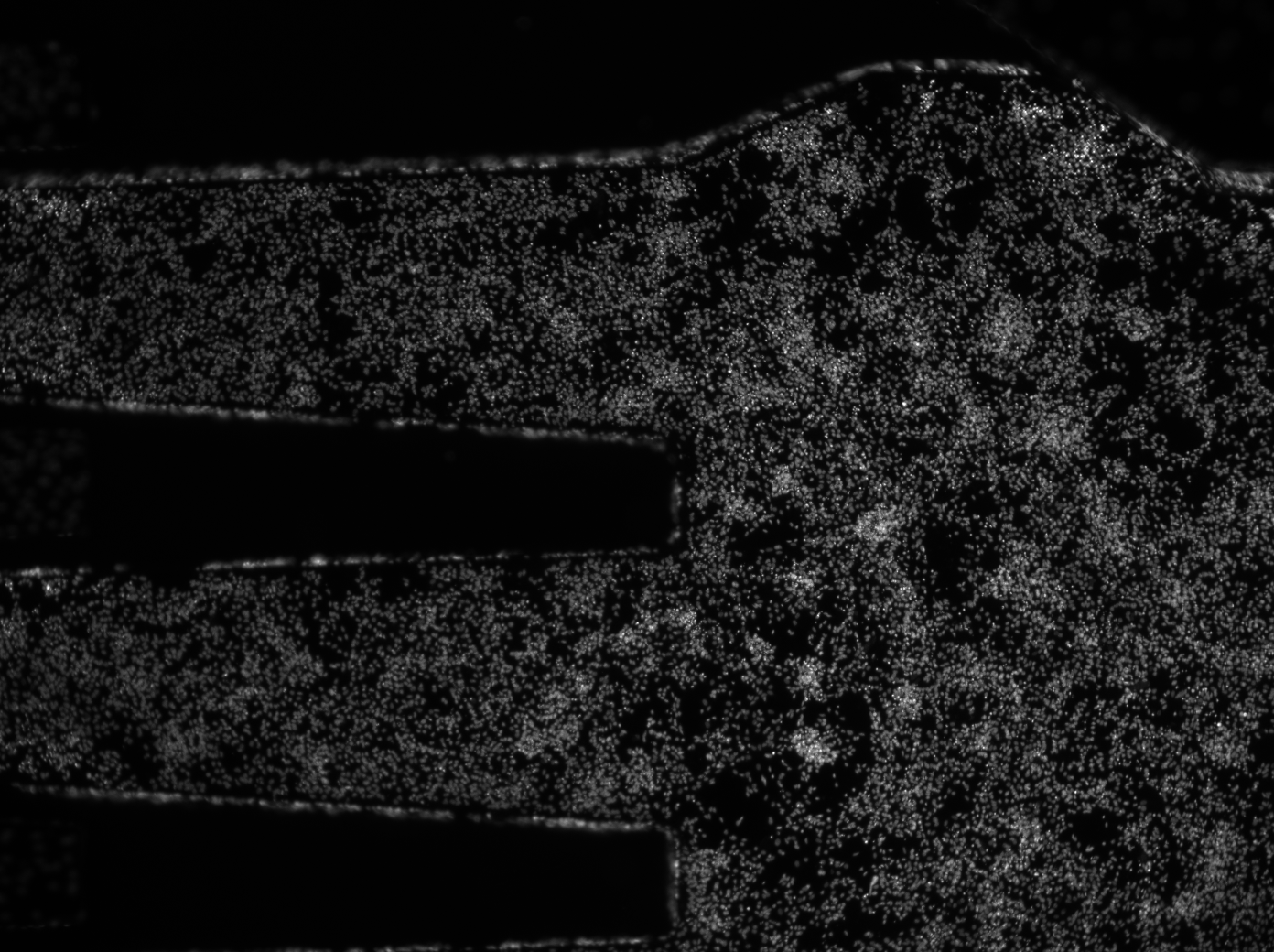 Hello all, I am trying to count the number of cells (white dots) and detect the x,y coordinates of each one in this image. I've tried using Fiji to do this but I can't detect all of the individual cells very well. This is what I've tried: 1. Process>>Filter>>Gaussian Blur to run a Gaussian Blur Filter (I've also tried without running this filter) 2. Image>>Adjust>>Threshold>>Select Dark Background to separate the cells from the background 3. Process>>Binary>>Watershed to try to separate touching cells, but it doesn't separate larger clumps of cells very well as shown below. 
Can I do any additional processing so that I can identify individual cells better? I don't need to preserve the size of the cells, just their position and the total number of cells. I would greatly appreciate help with this!!!! Thank you! Katrina |
Re: count nuclei in low mag image
|
You might want to try an adaptive threshold. Not 100% how to do it in
imageJ actually, but here's an example: http://scikit-image.org/docs/dev/auto_examples/plot_threshold_adaptive.html I'd expect this would clean up the contrast differences which are causing the larger images to be over-adjusted. On Thu, Nov 21, 2013 at 9:16 PM, katrina <[hidden email]> wrote: > <http://imagej.1557.x6.nabble.com/file/n5005647/Cocx_1_2.gif> > Hello all, > > I am trying to count the number of cells (white dots) and detect the x,y > coordinates of each one in this image. I've tried using Fiji to do this > but > I can't detect all of the individual cells very well. This is what I've > tried: > > 1. Process>>Filter>>Gaussian Blur to run a Gaussian Blur Filter (I've also > tried without running this filter) > 2. Image>>Adjust>>Threshold>>Select Dark Background to separate the cells > from the background > 3. Process>>Binary>>Watershed to try to separate touching cells, but it > doesn't separate larger clumps of cells very well as shown below. > <http://imagej.1557.x6.nabble.com/file/n5005647/Cocx_1_2_segmented.gif> > > Can I do any additional processing so that I can identify individual cells > better? I don't need to preserve the size of the cells, just their > position > and the total number of cells. > > I would greatly appreciate help with this!!!! Thank you! > > Katrina > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/count-nuclei-in-low-mag-image-tp5005647.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: count nuclei in low mag image
|
In reply to this post by katrina
Hi Katrina,
If you only need to know how many cells there are and where they are (but not size, intensity, etc.), then I would do this by using Process - Find Maxima. You can get the coordinates for each maxima plus total number of cells/nuclei. You can play around with the Noise tolerance if needed to make sure that you are getting the right selection and Preview point selection. You can add the selections to the ROI Manager and save them if you want to do that. You have choices for the Output type as well. I assume that your original image is larger than this but even this one worked well for me. Kind regards, Jacqui Jacqueline Ross Biomedical Imaging Microscopist Biomedical Imaging Research Unit School of Medical Sciences Faculty of Medical & Health Sciences The University of Auckland Private Bag 92019 Auckland 1142, NEW ZEALAND Tel: 64 9 923 7438 Fax: 64 9 373 7484 http://www.fmhs.auckland.ac.nz/sms/biru/ -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of katrina Sent: Friday, 22 November 2013 3:17 p.m. To: [hidden email] Subject: count nuclei in low mag image <http://imagej.1557.x6.nabble.com/file/n5005647/Cocx_1_2.gif> Hello all, I am trying to count the number of cells (white dots) and detect the x,y coordinates of each one in this image. I've tried using Fiji to do this but I can't detect all of the individual cells very well. This is what I've tried: 1. Process>>Filter>>Gaussian Blur to run a Gaussian Blur Filter (I've also tried without running this filter) 2. Image>>Adjust>>Threshold>>Select Dark Background to separate the cells from the background 3. Process>>Binary>>Watershed to try to separate touching cells, but it doesn't separate larger clumps of cells very well as shown below. <http://imagej.1557.x6.nabble.com/file/n5005647/Cocx_1_2_segmented.gif> Can I do any additional processing so that I can identify individual cells better? I don't need to preserve the size of the cells, just their position and the total number of cells. I would greatly appreciate help with this!!!! Thank you! Katrina -- View this message in context: http://imagej.1557.x6.nabble.com/count-nuclei-in-low-mag-image-tp5005647.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by katrina
Hi Katrina,
You may use the ITCN (Image-based Tool for Counting Nuclei) plugin. http://www.bioimage.ucsb.edu/automatic-nuclei-counter-plug-in-for-imagej Try the "width" from 6-8 pixels, "threshold" from 0.3-0.5, you could get various images with detected cells. chin |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

