segmentation problen
|
Dear all,
I am trying to accurately segment cells from immunostainings. Identifying cells is in a first trial based on staining nuclei. As can be seen in the accompanying image, the "nucleus counter" plugin of ImageJ performs reasonably well with most nuclei, and even with some which are touching. Nevertheless, there are many where neither he plugin nor any other way I have tried, succeeds in doing this satisfactorliy for real life images. Now one would imagine, that there is enough information in the pair of nuclei slightly off-centre towards 11:00 hours in the sample image, to identify it as two, like center of mass based on their non-touching outlines. Howver, my knowledge of how to use this information is more than limited. I would appreciate suggestions how to solve that problem. As the project advances, I certainly will stumble on more. Thanks very much. Matthias 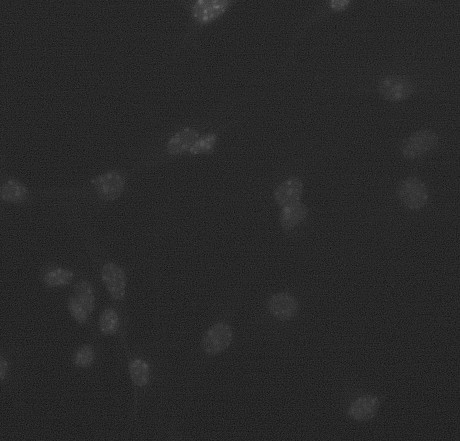
|
|
Hi Matthias Kirsch,
I have a hack that might work if you just want to count nuclei or identify double nuclei so you can blacklist them. Based on your image it looks like all the nuclei has uniform size. The one at 11:00 hours is almost twice as big. Find the median size of the nuclei and make a rule that say if the area of a particle is bigger than 1.5 times of this then it is a double nuclei. I have not tried the "nucleus counter" plugin of ImageJ, so I am not sure what format it will give you as output, but if you can export it to Excel, you can code this suggestion with a simple Excel macro. For a real solution you could look at the big particles. Trace their edge and look for 2 inflection points and draw a straight line between them. If that splits the particle roughly evenly this is a good candidate for splitting the nucleus. The ShapeLogic plugin has code for tracing the edge and finding inflection points, but this would involve some non trivial programming on your part. -Sami Badawi http://www.shapelogic.org On Fri, Oct 31, 2008 at 5:25 AM, Matthias Kirsch <[hidden email]> wrote: > Dear all, > > I am trying to accurately segment cells from immunostainings. Identifying > cells is in a first trial based on staining nuclei. As can be seen in the > accompanying image, the "nucleus counter" plugin of ImageJ performs > reasonably well with most nuclei, and even with some which are touching. > Nevertheless, there are many where neither he plugin nor any other way I > have tried, succeeds in doing this satisfactorliy for real life images. Now > one would imagine, that there is enough information in the pair of nuclei > slightly off-centre towards 11:00 hours in the sample image, to identify it > as two, like center of mass based on their non-touching outlines. Howver, my > knowledge of how to use this information is more than limited. > > I would appreciate suggestions how to solve that problem. As the project > advances, I certainly will stumble on more. > > Thanks very much. > > Matthias http://n2.nabble.com/file/n1402433/nuclei.jpg > -- > View this message in context: http://n2.nabble.com/segmentation-problen-tp1402433p1402433.html > Sent from the ImageJ mailing list archive at Nabble.com. > |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

