Re: object-oriented segmentation
Posted by Michiel van Dommelen on Dec 11, 2012; 2:29pm
URL: http://imagej.273.s1.nabble.com/object-oriented-segmentation-tp5001110p5001115.html
Dear Kees,
Thanks for the quick response. I am afraid this is not exactly what I am looking for. The way I interpreted your two methods, both “Vonoroi” as well as “Find Maxima” on the nuclear signal do not take the cytoplasmic signal into account. This way, the ROIs that are created are much bigger than the cytoplasmic signal. Or am I misinterpreting your advice?
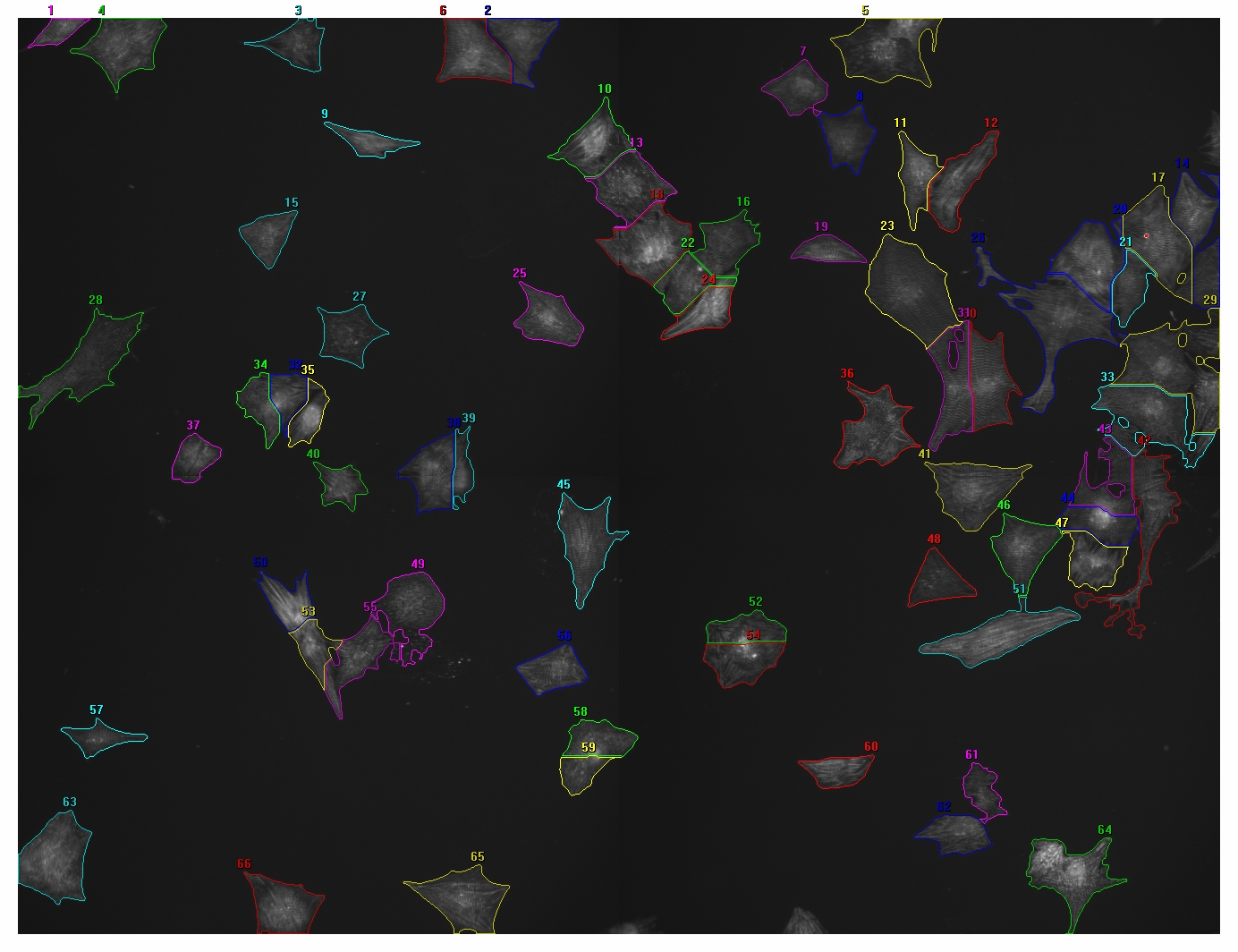
I will be a bit more explanatory in my question. This time I included two images, one with nuclear signal (Hoechst) and one with cytoplasmic signal (Actinin), processed with a different program. The segmentation is not perfect, but this is more or less what I would like to do with ImageJ.
Unfortunately not all of the cells have Actinin, on average about 10% of the nuclei do not have Actinin. I am only interested in the cells that do contain Actinin. Two of the parameters I am looking for are cellular area (either pixels or square um) and average intensity per cell. Eventually I would also like to transfer the ROIs to a third channel image for quantification of a third color. And ultimately I would like to automate this to batch process hundreds of images like this.
Is there a way to use “Vonoroi” or “Find Maxima” in such a way that the edges of the ROIs end with the edges of the cytoplasmic signal (Actinin)? Or is there a different way to obtain these ROIs?
Thanks again,
Michiel

URL: http://imagej.273.s1.nabble.com/object-oriented-segmentation-tp5001110p5001115.html
Dear Kees,
Thanks for the quick response. I am afraid this is not exactly what I am looking for. The way I interpreted your two methods, both “Vonoroi” as well as “Find Maxima” on the nuclear signal do not take the cytoplasmic signal into account. This way, the ROIs that are created are much bigger than the cytoplasmic signal. Or am I misinterpreting your advice?
I will be a bit more explanatory in my question. This time I included two images, one with nuclear signal (Hoechst) and one with cytoplasmic signal (Actinin), processed with a different program. The segmentation is not perfect, but this is more or less what I would like to do with ImageJ.
Unfortunately not all of the cells have Actinin, on average about 10% of the nuclei do not have Actinin. I am only interested in the cells that do contain Actinin. Two of the parameters I am looking for are cellular area (either pixels or square um) and average intensity per cell. Eventually I would also like to transfer the ROIs to a third channel image for quantification of a third color. And ultimately I would like to automate this to batch process hundreds of images like this.
Is there a way to use “Vonoroi” or “Find Maxima” in such a way that the edges of the ROIs end with the edges of the cytoplasmic signal (Actinin)? Or is there a different way to obtain these ROIs?
Thanks again,
Michiel


| Free forum by Nabble | Disable Popup Ads | Edit this page |