Grid Stitching - Can't OK image selection when >50 Images to stitch
Posted by asullivan on
URL: http://imagej.273.s1.nabble.com/Grid-Stitching-Can-t-OK-image-selection-when-50-Images-to-stitch-tp5018541.html
I'm having an issue stitching large numbers of pictures together with Grid Stitching. Basically the window that opens up for my approval is too large to fit on my screen and there is no scroll bar which would allow me scroll down to the end and process the images. I tried breaking up the images into stacks of 40 or so, but this became tedious.
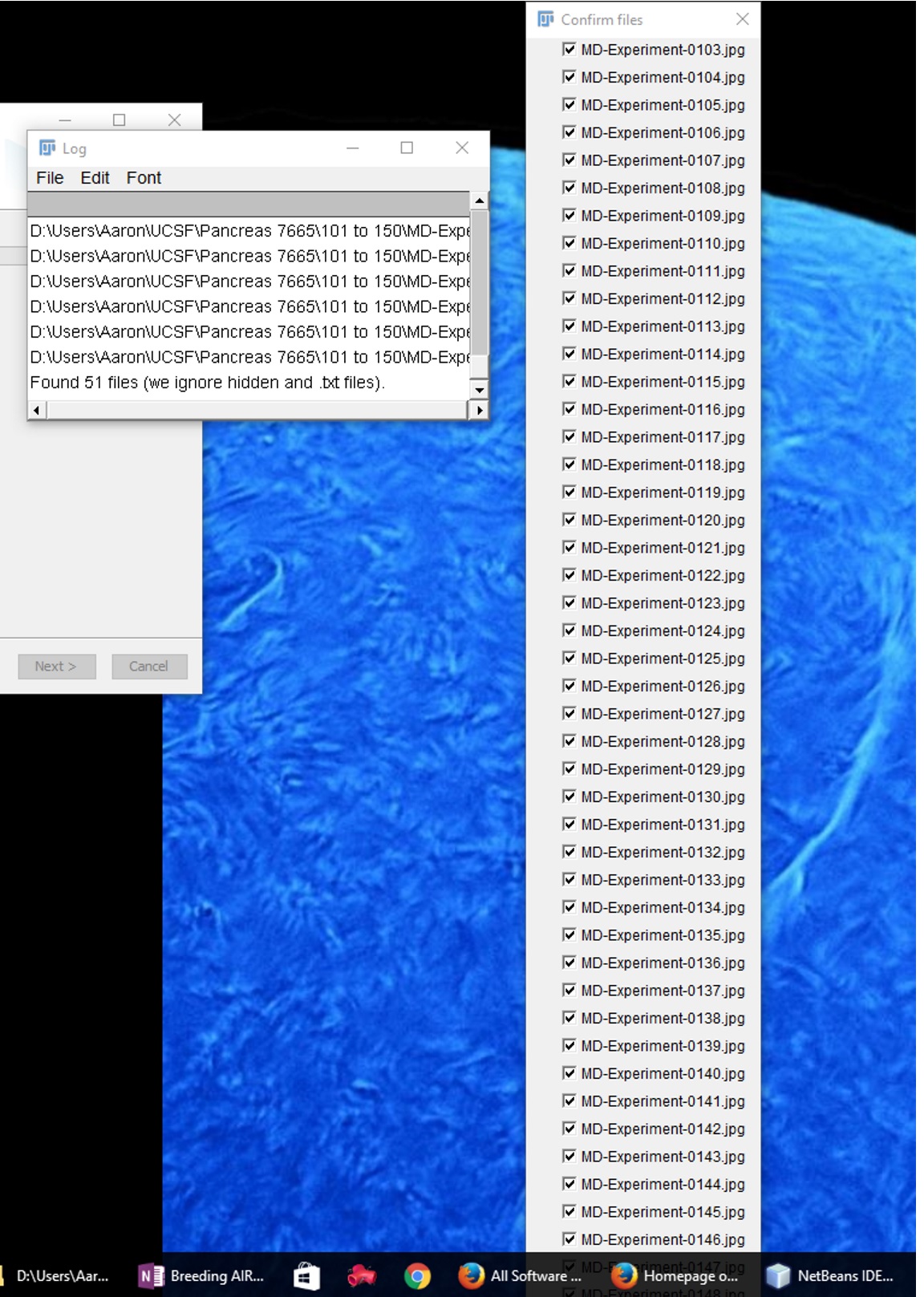
My second attempt to bypass this issue was to write a macro so it would enter in the files for me and I wouldn't have to hit OK.
My Macro
// Macro for stitching together lots of images.
inDir = getDirectory("Choose input directory"); //prompt user to choose input directory
outDir = getDirectory("Choose input directory"); // prompt usere to choose save/output directory
//Create an array of all files in inDir
arr = getFileList(inDir);
//Create a string of all file names separated by a space
str = "";
for (i=0; i<arr.length; i++) {
//Double check that file name is *.tif or *jpg
if(endsWith(arr[i], ".jpg")){
str += " " + arr[i];
}
if(endsWith(arr[i], ".tif")){
str += " " + arr[i];
}
}
inDir = replace(inDir, "\\\\$", " ");
run("Grid/Collection stitching", "type=[Unknown position] order=[All files in directory] directory=["+inDir+"] confirm_files output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 frame=1 ignore_z_stage display_fusion computation_parameters=[Save computation time (but use more RAM)] image_output=[Fuse and display] "+str+"");
But this didn't work. I am not able to process the images this way either.
Log File says this:
Stitching internal version: 1.2
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0001.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0002.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0005.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0006.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0010.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0011.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0012.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0013.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0014.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0015.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0016.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0017.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0018.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0019.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0021.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0022.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0023.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0024.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0025.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0026.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0027.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0028.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0029.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0030.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0031.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0032.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0033.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0034.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0035.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0036.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0037.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0038.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0039.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0040.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0041.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0042.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0043.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0044.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0045.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0046.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0047.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0048.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0049.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0050.jpg
Found 44 files (we ignore hidden and .txt files).
ERROR: Only 0 files selected, you need at least 2 - stop.
ERROR: Error during tile discovery, or invalid grid type. Aborting.
Would appreciate some help either with the plugin or with my macro.
URL: http://imagej.273.s1.nabble.com/Grid-Stitching-Can-t-OK-image-selection-when-50-Images-to-stitch-tp5018541.html
I'm having an issue stitching large numbers of pictures together with Grid Stitching. Basically the window that opens up for my approval is too large to fit on my screen and there is no scroll bar which would allow me scroll down to the end and process the images. I tried breaking up the images into stacks of 40 or so, but this became tedious.

My second attempt to bypass this issue was to write a macro so it would enter in the files for me and I wouldn't have to hit OK.
My Macro
// Macro for stitching together lots of images.
inDir = getDirectory("Choose input directory"); //prompt user to choose input directory
outDir = getDirectory("Choose input directory"); // prompt usere to choose save/output directory
//Create an array of all files in inDir
arr = getFileList(inDir);
//Create a string of all file names separated by a space
str = "";
for (i=0; i<arr.length; i++) {
//Double check that file name is *.tif or *jpg
if(endsWith(arr[i], ".jpg")){
str += " " + arr[i];
}
if(endsWith(arr[i], ".tif")){
str += " " + arr[i];
}
}
inDir = replace(inDir, "\\\\$", " ");
run("Grid/Collection stitching", "type=[Unknown position] order=[All files in directory] directory=["+inDir+"] confirm_files output_textfile_name=TileConfiguration.txt fusion_method=[Linear Blending] regression_threshold=0.30 max/avg_displacement_threshold=2.50 absolute_displacement_threshold=3.50 frame=1 ignore_z_stage display_fusion computation_parameters=[Save computation time (but use more RAM)] image_output=[Fuse and display] "+str+"");
But this didn't work. I am not able to process the images this way either.
Log File says this:
Stitching internal version: 1.2
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0001.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0002.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0005.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0006.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0010.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0011.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0012.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0013.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0014.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0015.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0016.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0017.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0018.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0019.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0021.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0022.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0023.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0024.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0025.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0026.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0027.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0028.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0029.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0030.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0031.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0032.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0033.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0034.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0035.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0036.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0037.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0038.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0039.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0040.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0041.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0042.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0043.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0044.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0045.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0046.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0047.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0048.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0049.jpg
D:\Users\Aaron\UCSF\Pancreas 7665\001 to 050\MD-Experiment-0050.jpg
Found 44 files (we ignore hidden and .txt files).
ERROR: Only 0 files selected, you need at least 2 - stop.
ERROR: Error during tile discovery, or invalid grid type. Aborting.
Would appreciate some help either with the plugin or with my macro.
| Free forum by Nabble | Edit this page |