Count foci within each cell
|
Hi,
I would like to count foci within each cell. Is there a way or macro to perform the approach? please have a look at the image. 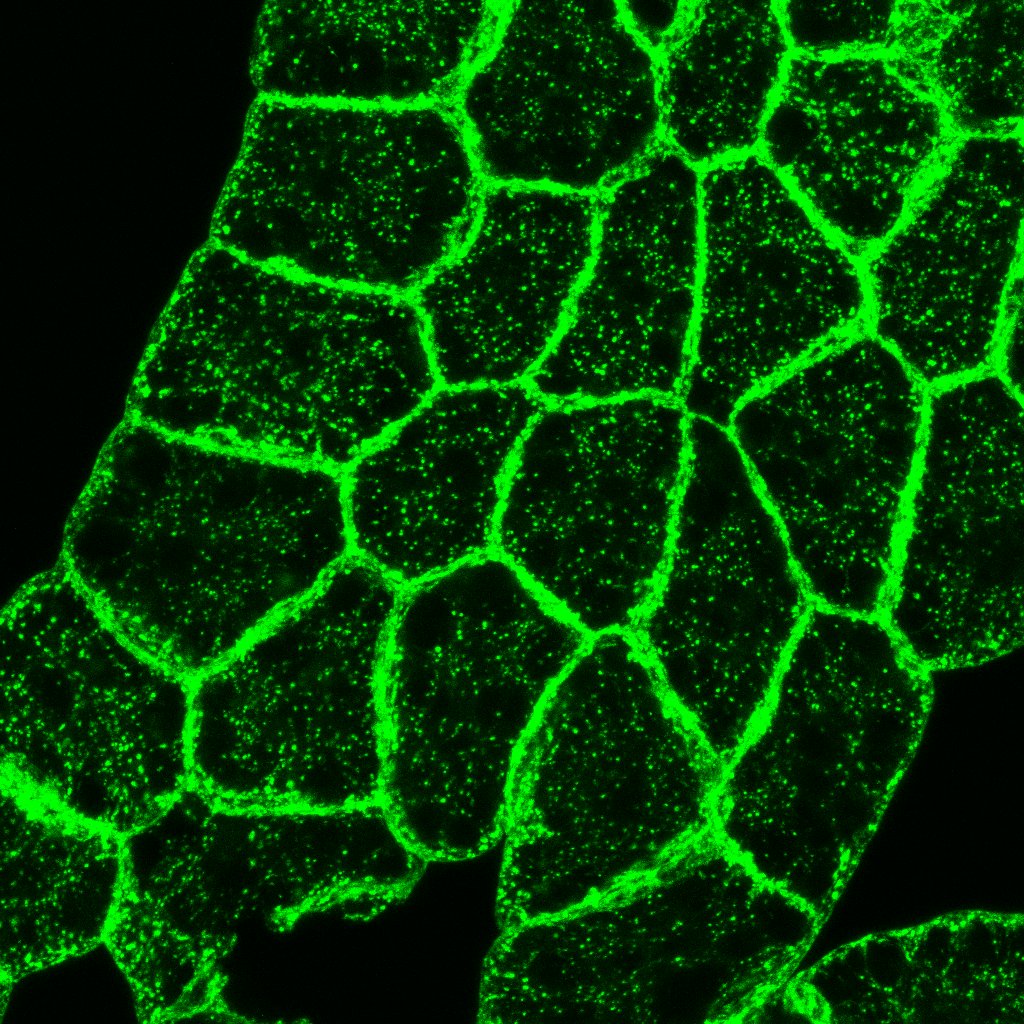 I tried to use threshold first but seem not obtain a good result. Any advice would be appreciated. Thanks in advance! chin |
|
On 3/4/14, 12:10 PM, chin wrote:
> Hi, > > I would like to count foci within each cell. Is there a way or macro to > perform the approach? > please have a look at the image. > <http://imagej.1557.x6.nabble.com/file/n5006766/C1foci..jpg> > I tried to use threshold first but seem not obtain a good result. Any advice > would be appreciated. > Try Process>Find Maxima... A noise tolerance of 20 looks ok. You can also find the borders of the cells by thresholding the image, and then discarding the small objects and closing the result. Gabriel Landini's BinaryFilterReconstruct is a good way to delete the foci while leaving the borders. --aryeh > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Count-foci-within-each-cell-tp5006766.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- Aryeh Weiss Faculty of Engineering Bar Ilan University Ramat Gan 52900 Israel Ph: 972-3-5317638 FAX: 972-3-7384051 -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by chin
Hi Chin,
I outlined a possible procedure regarding your sample image. This might need some adjustments for different images //for the cellular outlines: run("Subtract Background...", "rolling=20"); run("Median...", "radius=8"); run("Enhance Contrast...", "saturated=0 normalize"); setAutoThreshold("Huang dark"); //this was a quick and dirty trial, different setOption("BlackBackground", true); run("Convert to Mask"); run("Invert"); run("Fill Holes"); //run the "Watershed Irregular Features" macro from the BioVoxxel Toolbox (e.g. with an erosion cycle of 8 or manually close areas of cells according to the original image if not possible differently) // for extracting the small spots run("Subtract Background...", "rolling=20"); run("Enhance Contrast...", "saturated=0 normalize"); //run the "Median Background Subtraction" from the BioVoxxel Toolbox with a dilation of 3 and a median filter radius of 10 pixels setAutoThreshold("IsoData dark"); //or a threshold which performes better in this casesetOption("BlackBackground", true); setOption("BlackBackground", true); run("Convert to Mask"); run("Watershed"); //run the "Speckle Inspector" from the BioVoxxel Toolbox //There identify the two different images and your parameters if necessary //see also: http://fiji.sc/BioVoxxel_Toolbox#Speckle_Inspector [image: Inline-Bild 3] Hope this helps a little with your analysis. kind regards, Jan 2014-03-04 11:10 GMT+01:00 chin <[hidden email]>: > Hi, > > I would like to count foci within each cell. Is there a way or macro to > perform the approach? > please have a look at the image. > <http://imagej.1557.x6.nabble.com/file/n5006766/C1foci..jpg> > I tried to use threshold first but seem not obtain a good result. Any > advice > would be appreciated. > > Thanks in advance! > > chin > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Count-foci-within-each-cell-tp5006766.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Thanks both aryeh and Jan's suggestion.
I could almost figure out and learn the steps from it. However, i am still confused a little bit about the those folloing steps: From aryeh's method: //You can also find the borders of the cells by thresholding the image, and then discarding the small objects and closing the result. // From Jan's method: //run the "Watershed Irregular Features" macro from the BioVoxxel Toolbox (e.g. with an erosion cycle of 8 or manually close areas of cells according to the original image if not possible differently) How could I "manually close areas of cells according to the original image if not possible differently" Could you explain it more detail? Thanks again. Best regard, chin |
|
On 3/6/14, 2:57 AM, chin wrote:
> Thanks both aryeh and Jan's suggestion. > > I could almost figure out and learn the steps from it. > However, i am still confused a little bit about the those folloing steps: > > >From aryeh's method: > //You can also find the borders of the cells by thresholding the image, > and then discarding the small objects and closing the result. // > ok -- here is the output of the macro recorder for a set of steps which detect the cell boundaries. // the following just grabs the image from the clipboard and extracts the green channel. run("System Clipboard"); run("Split Channels"); selectWindow("Clipboard (blue)"); close(); selectWindow("Clipboard (red)"); close(); selectWindow("Clipboard (green)"); //threshold the image -- Huang tends to set a lower threshold setAutoThreshold("Huang dark"); //run("Threshold..."); // the size option in the following line is what eliminates the small objects, // while selecting the cell boundaries run("Analyze Particles...", "size=10000-Infinity show=Masks add"); run("Grays"); //because the mask is created with an inverting LUT // the close operation gives a more continuous network. setOption("BlackBackground", true); run("Options...", "iterations=4 count=1 black edm=Overwrite do=Close"); You might also consider skeletonize and then prune to get cleaner boundaries. This will allow you count the foci in each cell separately. -- Aryeh Weiss Faculty of Engineering Bar Ilan University Ramat Gan 52900 Israel Ph: 972-3-5317638 FAX: 972-3-7384051 -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

