Hi all,
I'm following the protocol outlined here:
http://microscopy.duke.edu/HOWTO/countfoci.htmlAnd the commands I'm using are enclosed below.
My problem is that I'm working on massive images (25k x 17k pixels) and sometimes I get accurate values but sometimes I get what I see below. Going by results table (and by RawIntDen/255 to get the actual number of nuclear foci), the cluster highlighted has around 60,000 nuclei! ...which is not the case as you can see by a quick visual examination.
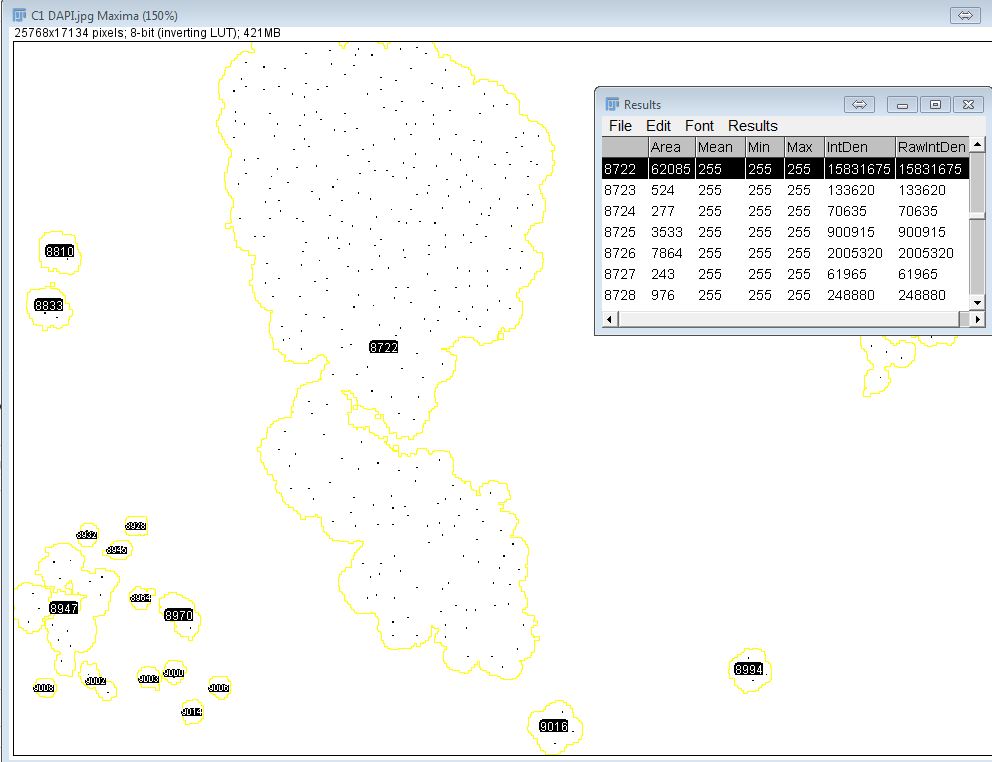
Any idea why this is or what I can do about it?
I've tried re-binarizing the image in the step immediately before measuring. That didn't work. I get this problem whether I do the commands manually or whether I do it via the macro. Any other ideas?
Thanks in advance.
run("8-bit");
run("Auto Threshold", "method=Triangle white setthreshold show");
run("Convert to Mask");
run("Fill Holes");
run("Dilate");
run("Dilate");
run("Analyze Particles...", "size=50-Infinity display clear summarize add in_situ");
wait(30000);
run("Revert");
run("Find Maxima...", "noise=7 output=[Single Points] exclude");
run("ROI Manager...");
roiManager("Show None");
roiManager("Show All");
run("Set Measurements...", "area mean min integrated redirect=None decimal=3");
roiManager("Measure");
wait(30000);
roiManager("Save", path + ".zip");
saveAs("Results", path + ".xls");
close();