Detecting dark spots (dead cells)
Detecting dark spots (dead cells)
|
CONTENTS DELETED
The author has deleted this message.
|
Re: Detecting dark spots (dead cells)
|
Hi Tom,
Well, the essential problems with this image are (1) the illumination is uneven, so that the background intensity varies from a pixel value of 110 to 130 as you traverse the image. (2) the intensity of the dark spots is very close to that of the background --i.e. 122 vs a background of 125. This is a very small differential. In general, the best solution for a problem like this is to get a better image. What would happen to the image if you used a red filter? That might darken the TP cells significantly compared to the background. Alternatively, you might be able to brighten the background to emphasize the dark cells. Joel On Mon, Jan 9, 2012 at 4:27 AM, Tom Runia <[hidden email]> wrote: > Hello everybody, > > I am trying to detect the dead cells in images such as these: > http://tweakers.net/ext/f/Zk9bURFkJoqlMAudOQwGLvY9/full.png > The dead cells are yeast cells marked with Trypan blue. My goal is to > determine the viability from this image and therefore I need to count all > the dead cells. > > My question is whether anybody has an idea about how to detect these dark > spots? I thought about finding the local minima and then checking the > surrounding pixels for each minima to see if the close surrounding is dark. > I tried to work this out, but the result is not very good, see the marked > points in this image: > http://tweakers.net/ext/f/Nz7UMpldsWcxBDhDND8id9qX/full.jpg > > Does anyone have a clever idea about how to detect these dark spots? Thanks > in advance!! > > Best, > > Tom > -- Joel B. Sheffield, Ph.D Department of Biology Temple University Philadelphia, PA 19122 Voice: 215 204 8839 e-mail: [hidden email] URL: http://astro.temple.edu/~jbs |
Re: Detecting dark spots (dead cells)
|
CONTENTS DELETED
The author has deleted this message.
|
Re: Detecting dark spots (dead cells)
|
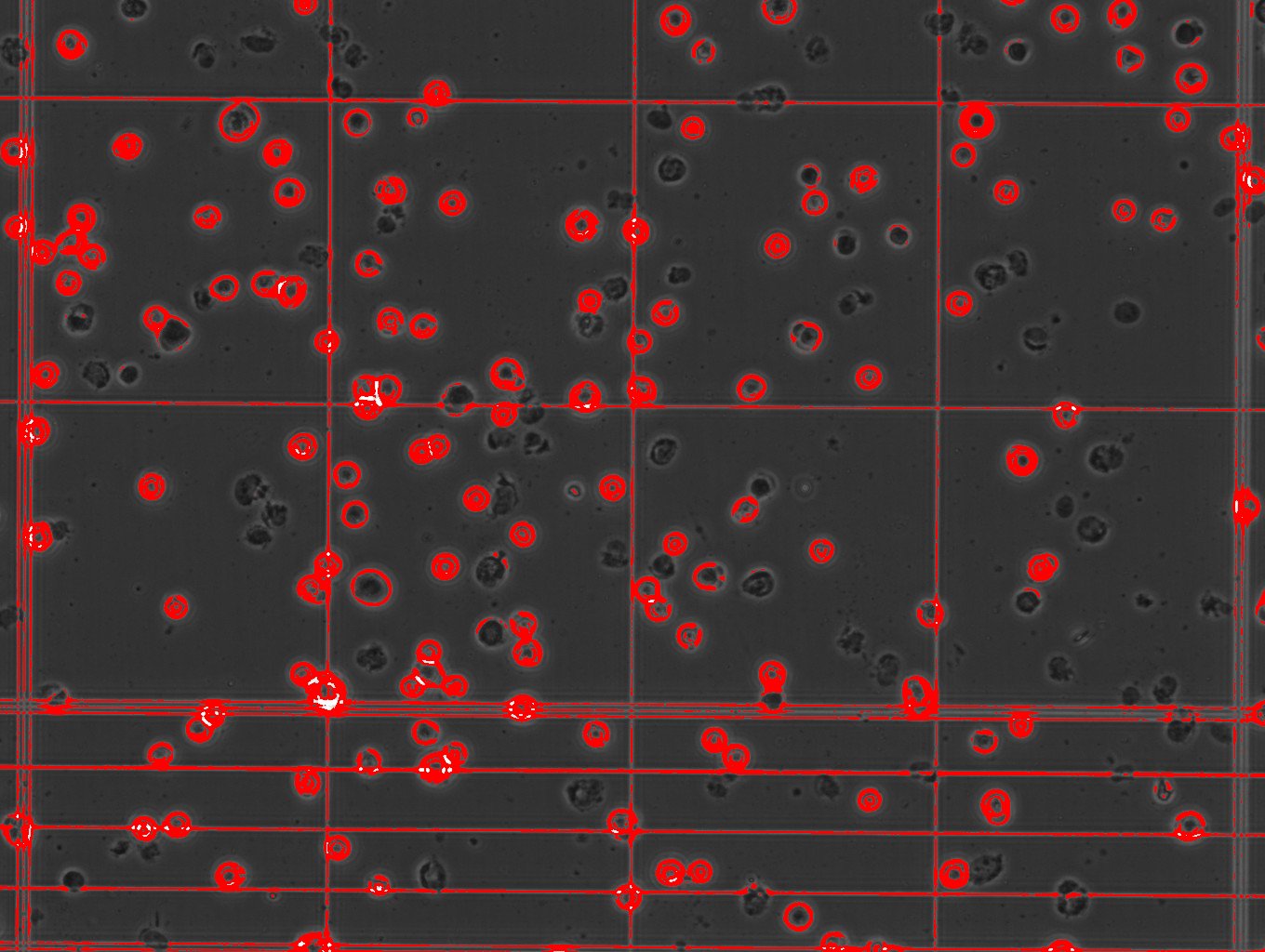
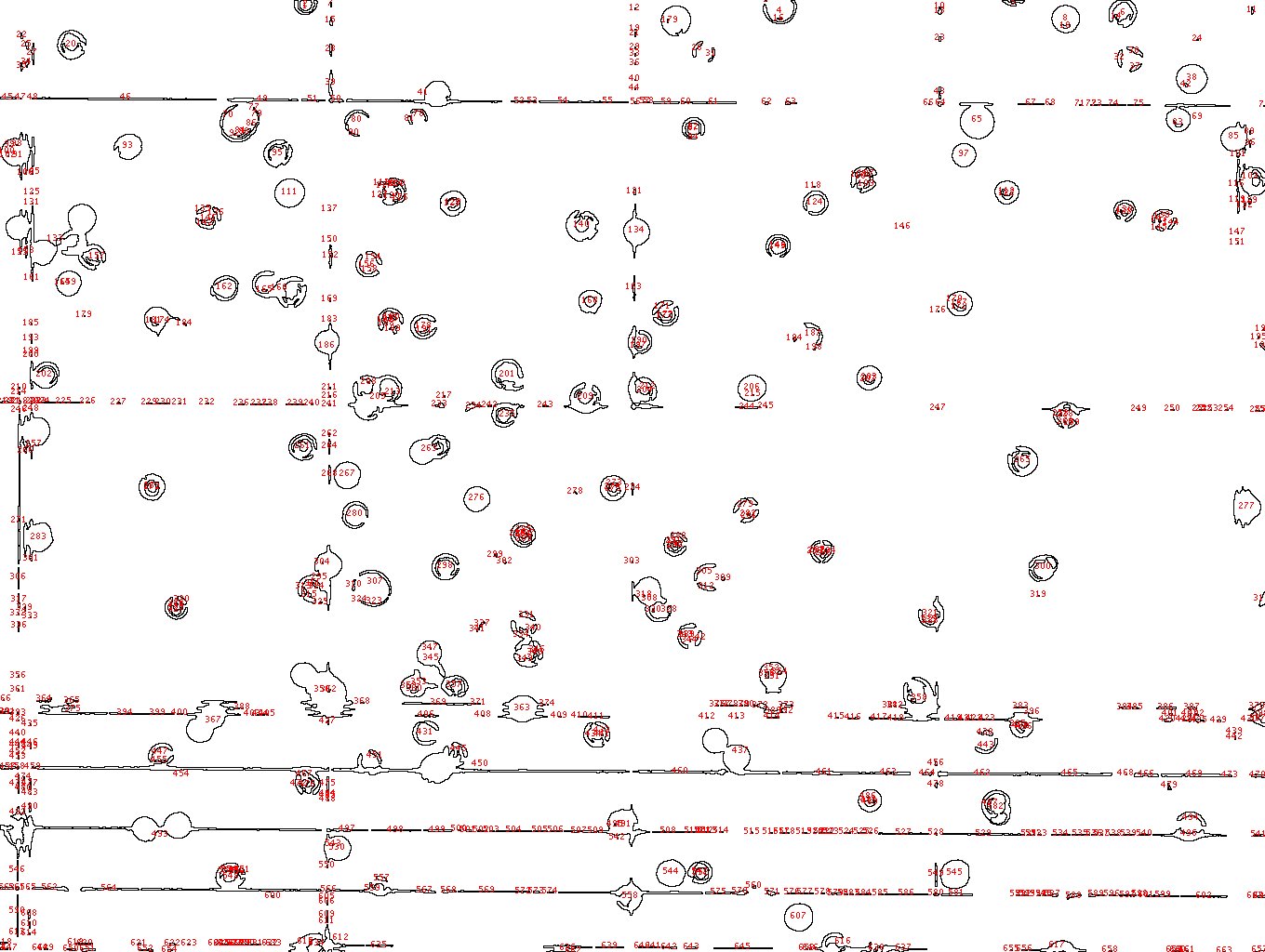
I'm having a similar issue, the resolution in my case is fine, I can visible discriminate bitten live and death cells as showed in the link
/Users/tizianarossetti/Desktop/cells viability.jpg the problem is with the analysis I have many quantification of the background that shouldn't be there. /Users/tizianarossetti/Desktop/cells counting analysis.jpg any suggestion? Cheers Tizy |
Re: Detecting dark spots (dead cells)
|
Re: Detecting dark spots (dead cells)
|
In reply to this post by Joel Sheffield
I'm having a similar issue, the resolution in my case is fine, I can visible discriminate bitten live and death cells as showed in the link
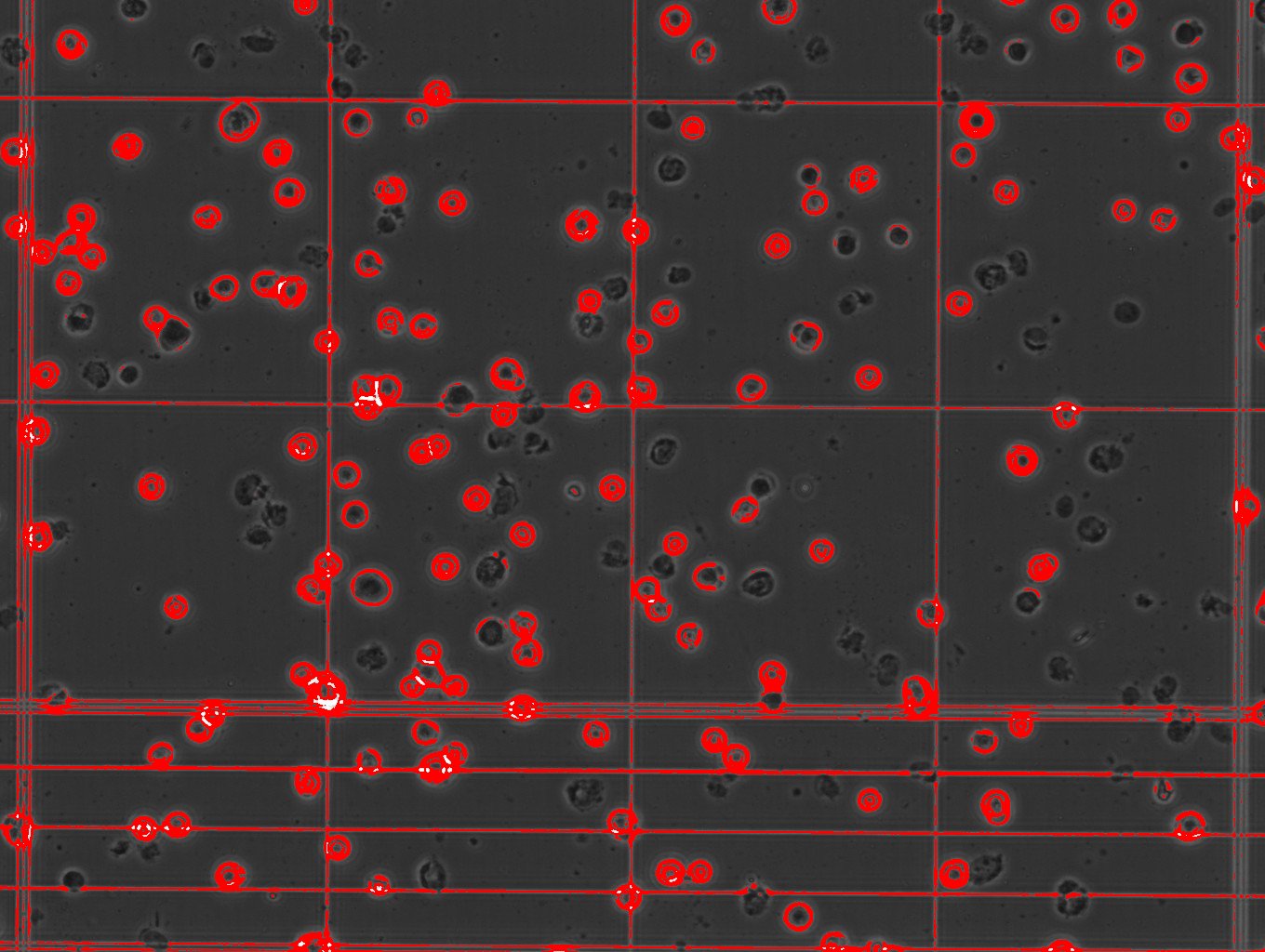 the problem is with the analysis I have many quantification of the background that shouldn't be there. 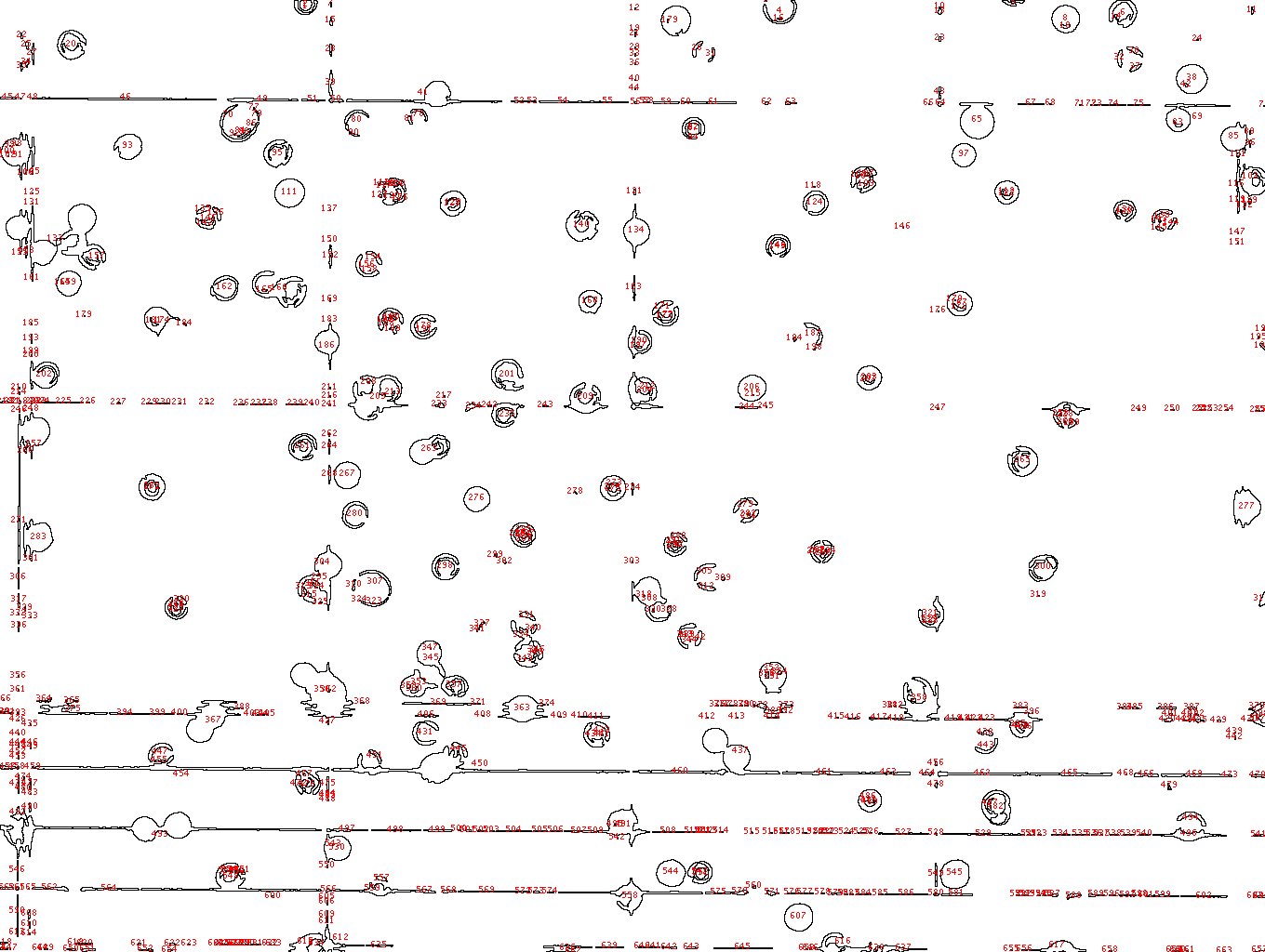 any suggestion? Cheers Tizy |
Re: Detecting dark spots (dead cells)
|
Hi Tizy,
as far as I can remember something like this has been on the list a while ago - the final solution was simply to avoid the grid. You need to calibrate the scale of your image separately (with your grid or any other object of known size) and keep the magnification. Michael ________________________________________________________________ On May 17, 2012, at 21:20, tizianarossetti wrote: > I'm having a similar issue, the resolution in my case is fine, I can visible > discriminate bitten live and death cells as showed in the link > > http://imagej.1557.n6.nabble.com/file/n4992111/cells_viability.jpg > > the problem is with the analysis I have many quantification of the > background that shouldn't be there. > > http://imagej.1557.n6.nabble.com/file/n4992111/cells_counting_analysis.jpg > > > any suggestion? > > Cheers > > Tizy |
Re: Detecting dark spots (dead cells)
|
Hi all,
I haven't found the older post mentionned, so I figured I'd just post this in reply. Unfortunately, it is not just about not using the grid. Cell counting chambers are specifically made to count cells in a volume (As in, there is a precise separation between the coverslip and the chamber - Usually 0.1mm) which allows you to do the counting accurately, even if you pipet a semi-variable amount each time. So if you want to use automatic methods, you still need a cell counting chamber-like device. It's not enough to put liquid on a glass slide and put a coverslip on it, the volumes would not match and probably be much less accurate. Even if you manage to get a counting chamber-like device without the grid, you still need to keep the stereology counting rules like for the manual counting (Keep objects if they intersect the bottom and left corners, discard if they are in contact with the upper and left corners) in your counting pipeline, or you rist over/under estimating the amount by a fair percentage. All in all, you risk having a less accurate count by doing it automatically than if you counted a few chambers manually... And loose more time in the acquisition and processing then if you had just counted them by hand. Best Oli Hi Tizy, as far as I can remember something like this has been on the list a while ago - the final solution was simply to avoid the grid. You need to calibrate the scale of your image separately (with your grid or any other object of known size) and keep the magnification. Michael ________________________________________________________________ On May 17, 2012, at 21:20, tizianarossetti wrote: > I'm having a similar issue, the resolution in my case is fine, I can visible > discriminate bitten live and death cells as showed in the link > > http://imagej.1557.n6.nabble.com/file/n4992111/cells_viability.jpg > > the problem is with the analysis I have many quantification of the > background that shouldn't be there. > > http://imagej.1557.n6.nabble.com/file/n4992111/cells_counting_analysis.jpg > > > any suggestion? > > Cheers > > Tizy |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

