Help measuring migration through neurospheres!
|
Hello all,
I'm having some issues figuring out an easier way to measure migration via my neurosphere assay. Essentially, for my assay, I form neurospheres by plating neural stem cells in absence of any coating substrate. Without coating, the neural stem cells free float in the media and over time aggregate to form spheres known as neurospheres. Once spheres are formed in the suspension, I collect and replate them onto dishes that are coated. The coating allows for the neurospheres to stick and for cells to migrate out from the original sphere. An example of what this looks like can be seen below. 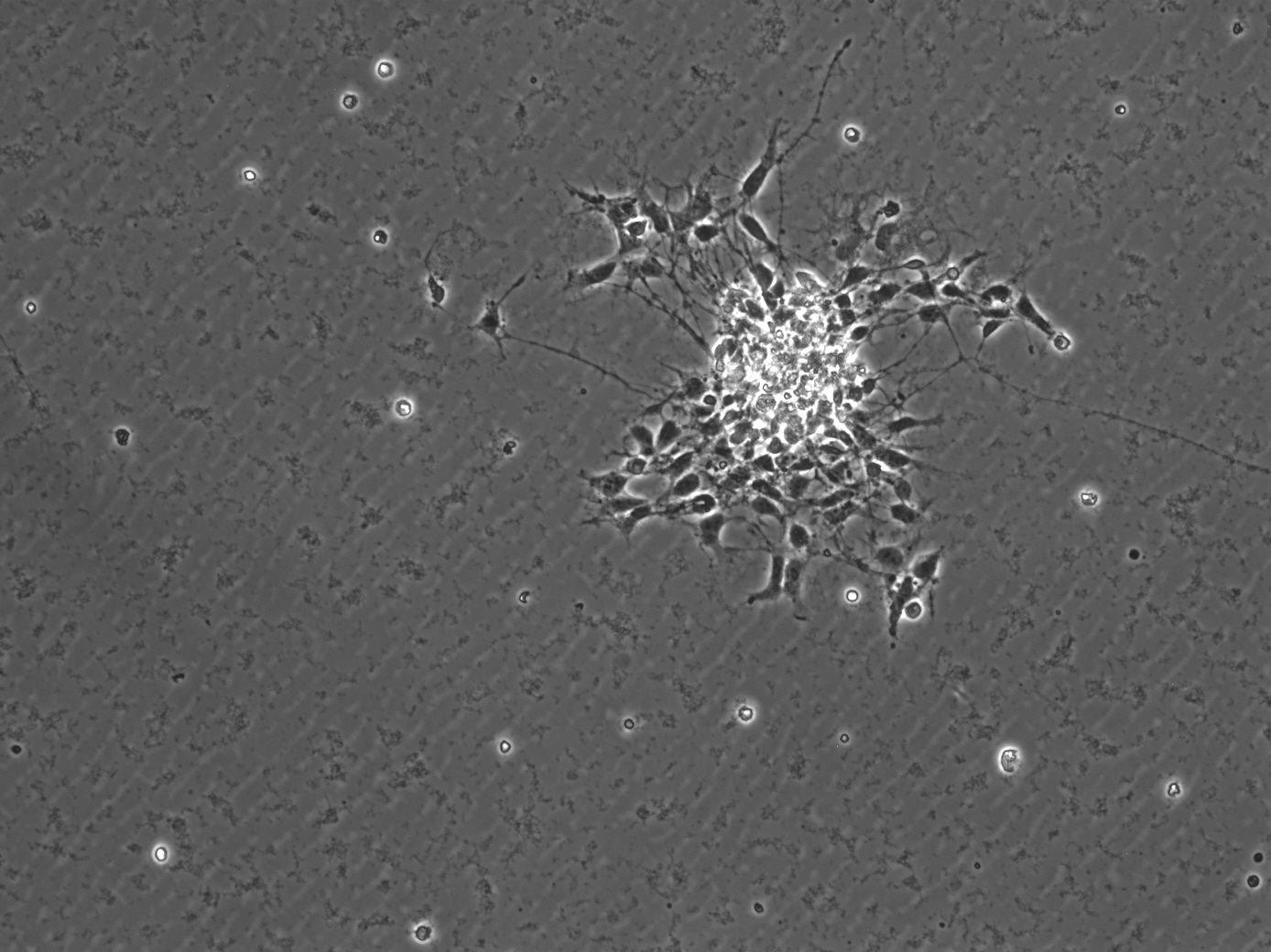 In order to quantify migration, I currently measure inner area of the sphere (which is the bright center) and subtract it from total area of the sphere. My measurements are done by manually tracing both the inner and outer sphere in image j using the free hand tool. I trace around all the outer cells and essentially trace a circle around the inner mass However, as I acquire more spheres this is becoming very time consuming! Does anyone know of an easier way to do this???? In order to quantify migration, I currently measure inner area of the sphere (which is the bright center) and subtract it from total area of the sphere. My measurements are done by manually tracing both the inner and outer sphere in image j using the free hand tool. I trace around all the outer cells and essentially trace a circle around the inner mass However, as I acquire more spheres this is becoming very time consuming! Does anyone know of an easier way to do this????
|
Re: Help measuring migration through neurospheres!
|
Hi smrpre,
you will find below a small macro that could be a starting point. It's far from being "perfect", but it could give you relatively "good" approximation. Up to now it's just measure area. thus (dead / round cells) around your sphere are also counted. But you could easily add an "analyse particles" to clean them all if you wish. There is a couple of parameters you can tune, size of the gaussian blur, threshold methods ... Copy and paste the code below in a macro editor open your image press run Regards, Romain ////////////////////////////////////////////////////////////////////////////// start of the macro userSelection = false ; ////////////////////////////////////////////////////////////////////////////// set the measurements run("Set Measurements...", "area limit display redirect=None decimal=3"); // area, limited to threshold, display label title = getTitle(); run("8-bit"); // the image is RGB, convert it to 8-bit because it's grey anyway ////////////////////////////////////////////////////////////////////////////// detect the inner core innerImageName = title+"-inner"; run("Duplicate...", "title="+innerImageName); // duplicate the image with an explicite name run("Gaussian Blur...", "sigma=20"); // blur it run("Threshold..."); // threshold setAutoThreshold("Default dark"); // intermodes gives also a good approximation if (userSelection){ // if you declare userSelection = true (line 1), waitForUser("Please Select a threshold"); // you can select a threshold, } // (should be the same for all the images zou would like to compare) // run("Measure"); // measure the delimited area setOption("BlackBackground", true); // make a binary image of it run("Convert to Mask"); // ////////////////////////////////////////////////////////////////////////////// detect the sprout selectImage(title); run("FeatureJ Edges", "compute smoothing=1.0 lower=[] higher=[]"); // detect the edges to detect the cells contours and protrusion setAutoThreshold("Li dark"); // threshold setOption("BlackBackground", true); // run("Convert to Mask"); // run("Gaussian Blur...", "sigma=5"); // blur it, because there is a lot of artifacts (coating agggregates ?) setAutoThreshold("Default dark"); // threshold again, Yen is a bit more permissive expandImageName = title+"-sprout"; rename(expandImageName); if (userSelection){ // if you declare userSelection = true (line 1), waitForUser("Please Select a threshold"); // you can select a threshold, } // (should be the same for all the images zou would like to compare) run("Measure"); // measure the delimited area setOption("BlackBackground", true); // make a binary image of it run("Convert to Mask"); // /////////////////////////////////////////////////////////////////////////////// create an output image run("Merge Channels...", "c1=["+innerImageName+"] c2=["+expandImageName+"]");// merge the two mask selectImage("RGB"); rename(title+"_output"); // rename it run("Add Image...", "image=["+title+"] x=0 y=0 opacity=80"); // add the original image as an overlay ////////////////////////////////////////////////////////////////////////////// end of the macro --------------------------------------------------------------- Dr. Romain Guiet Bioimaging and Optics Platform (PT-BIOP) Ecole Polytechnique Fédérale de Lausanne (EPFL) Faculty of Life Sciences Station 19, AI 0140 CH-1015 Lausanne Phone: [+4121 69] 39629 http://biop.epfl.ch/ --------------------------------------------------------------- ________________________________________ De : ImageJ Interest Group [[hidden email]] de la part de smrpre [[hidden email]] Envoyé : jeudi 17 septembre 2015 18:49 À : [hidden email] Objet : Help measuring migration through neurospheres! Hello all, I'm having some issues figuring out an easier way to measure migration via my neurosphere assay. Essentially, for my assay, I form neurospheres by plating neural stem cells in absence of any coating substrate. Without coating, the neural stem cells free float in the media and over time aggregate to form spheres known as neurospheres. Once spheres are formed in the suspension, I collect and replate them onto dishes that are coated. The coating allows for the neurospheres to stick and for cells to migrate out from the original sphere. An example of what this looks like can be seen below. <http://imagej.1557.x6.nabble.com/file/n5014350/1_%281%29.jpg> In order to quantify migration, I currently measure inner area of the sphere (which is the bright center) and subtract it from total area of the sphere. My measurements are done by manually tracing both the inner and outer sphere in image j using the free hand tool. I trace around all the outer cells and essentially trace a circle around the inner mass However, as I acquire more spheres this is becoming very time consuming! Does anyone know of an easier way to do this???? -- View this message in context: http://imagej.1557.x6.nabble.com/Help-measuring-migration-through-neurospheres-tp5014350.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Thank you so much for this amazing macro. I am tweaking some of the parameters but it's working beautifully! On Oct 8, 2015 10:38 AM, "Romain Guiet [via ImageJ]" <[hidden email]> wrote:
Hi smrpre, |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

