How can I exclude the rounded (dead) cells in threshold measurement?
|
I am using the imagej to analyse Glucose uptake with a florescent derivative of Glucose(2-NBDG) in some epithelial and endothelial cells. I am using threshold and Integrated density for all the cells in the field of view (Image: Type->8bit, Adjust-> threshold, Analyze-> analyze Particle, IntDen). Since I have some dead roundish cells which show a false too bright florescent together with the alive spindle cells, attached to the well, I need to somehow exclude or mask these roundish cells to not affect the analysis. It is important for me to do the threshold measurement for all the cells, since I have dozen of pics per condition. I would be thankful if anyone can give me tips how I can filter or mask these ''too bright dead cells'' from the analysis of my data.
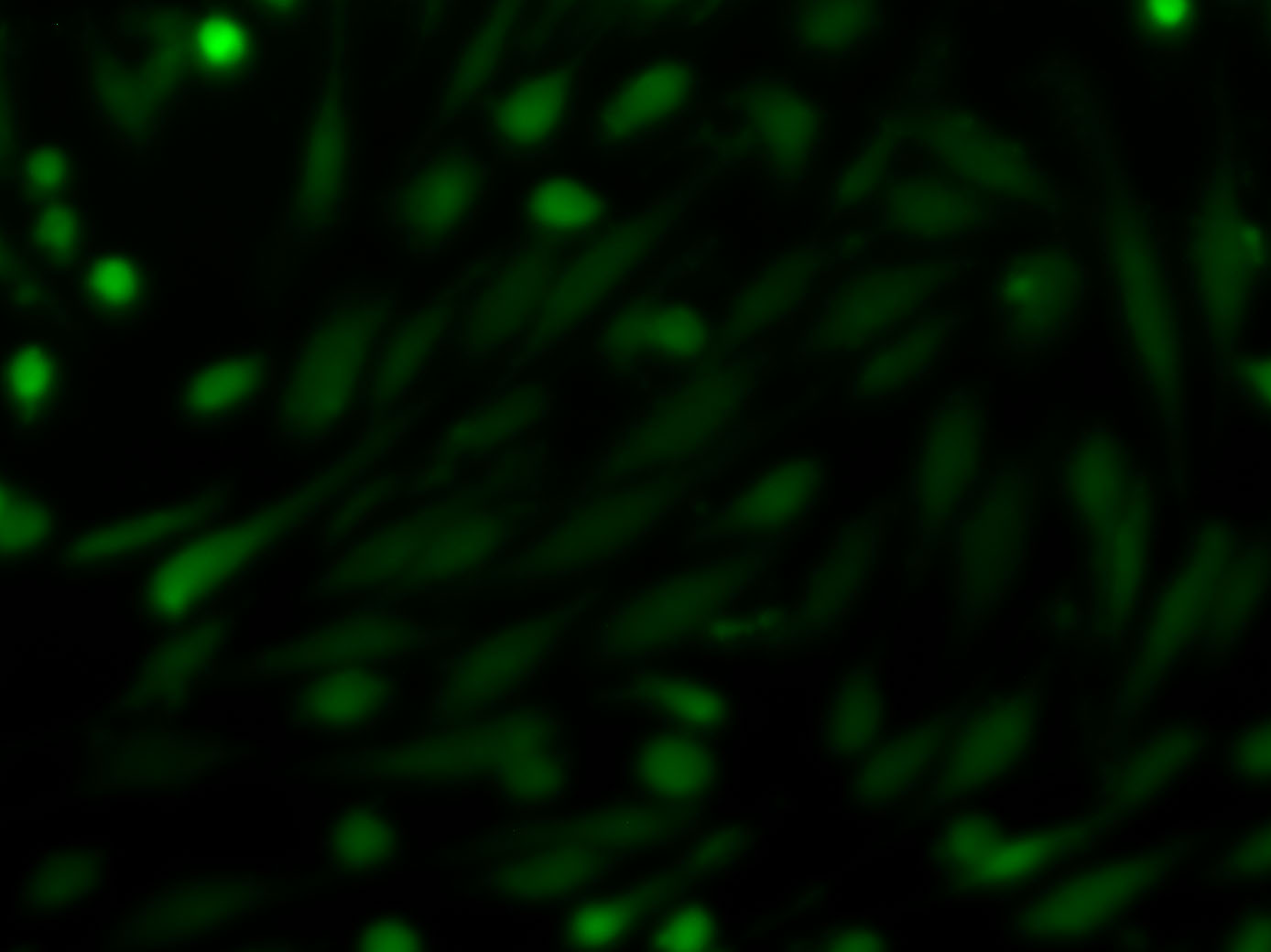 Bests, Azadeh |
|
Dear Azadeh,
Can you use in analyse particles the option "Circularity"? If you set this to 0.0 - 0.7 or something like you might be able to exclude the rounded cells. Best wishes Kees -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Azi.Nil Sent: 21 May 2013 10:47 To: [hidden email] Subject: How can I exclude the rounded (dead) cells in threshold measurement? I am using the imagej to analyse Glucose uptake with a florescent derivative of Glucose(2-NBDG) in some epithelial and endothelial cells. I am using threshold and Integrated density for all the cells in the field of view (Image: Type->8bit, Adjust-> threshold, Analyze-> analyze Particle, IntDen). Since I have some dead roundish cells which show a false too bright florescent together with the alive spindle cells, attached to the well, I need to somehow exclude or mask these roundish cells to not affect the analysis. It is important for me to do the threshold measurement for all the cells, since I have dozen of pics per condition. I would be thankful if anyone can give me tips how I can filter or mask these ''too bright dead cells'' from the analysis of my data. <http://imagej.1557.x6.nabble.com/file/n5003061/1125-260.jpg> Bests, Azadeh -- View this message in context: http://imagej.1557.x6.nabble.com/How-can-I-exclude-the-rounded-dead-cells-in-threshold-measurement-tp5003061.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Dear Kees,
Thanks for your suggestion! I found the circularity option and changed it to what u suggested. But could you pleas tell me how I can measure the circularity and what does this 0.00-0.7 mean? Bests, Azadeh |
|
Hi Azadeh,
On May 21, 2013, at 7:31 AM, Azi.Nil wrote: > Dear Kees, > > Thanks for your suggestion! I found the circularity option and changed it to > what u suggested. But could you pleas tell me how I can measure the > circularity and what does this 0.00-0.7 mean? Circularity is documented here... http://rsb.info.nih.gov/ij/docs/guide/146-30.html#toc-Subsection-30.7 In a nutshell, the closer circularity gets to the value 1 the more circular the object. Many of your cells are elongated so they will have low circularity values. Cheers, Ben P.S. Don't forget to include a bit of meaningful quote from the when replying to this list. Many of us don't use Nabble so we lose that context when Nabble users reply. > > Bests, > Azadeh > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/How-can-I-exclude-the-rounded-dead-cells-in-threshold-measurement-tp5003061p5003066.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Azi.Nil
Hi Azadeh,
Additionally, to the suggestions below and/or if circularity would not satisfy you completely as an exclusion criteria, you can also exclude cells/features by using the advanced particle analyzer from the BioVoxxel Toolbox under: http://fiji.sc/List_of_update_sites which gives you the flexibility to use further shape descriptors as determinants for the analysis. On the respective page is also the instruction how to include these sites in Fiji. Otherwise you can simply download the plain text from the respective macro toolset and save it as .txt-file in the >Fiji.app >macros >toolsets folder. best, Jan On May 21, 2013, at 7:31 AM, Azi.Nil wrote: > Dear Kees, > > Thanks for your suggestion! I found the circularity option and changed it to > what u suggested. But could you pleas tell me how I can measure the > circularity and what does this 0.00-0.7 mean? Circularity is documented here... http://rsb.info.nih.gov/ij/docs/guide/146-30.html#toc-Subsection-30.7 In a nutshell, the closer circularity gets to the value 1 the more circular the object. Many of your cells are elongated so they will have low circularity values. Cheers, Ben P.S. Don't forget to include a bit of meaningful quote from the when replying to this list. Many of us don't use Nabble so we lose that context when Nabble users reply. > > Bests, > Azadeh -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

