Index of hexagonality? [SOLVED]
|
This post was updated on .
Hi all,
I'm looking at some endothelial cells which are meant to be hexagonal and tightly packed in nature. With stress and/or trauma, these cells exhbit polymorphism - they turn into polygons of varying types. My aim is to determine how many cells have lost their hexagon shapes in order to infere the severity of the insult. I've attached two samples. The first is simply the cellls. The second has a yellow box to highlight a Hexagon whilst the red box highlights a cell that has undergone transformation into a a different polygon (pentagon - in this instance). 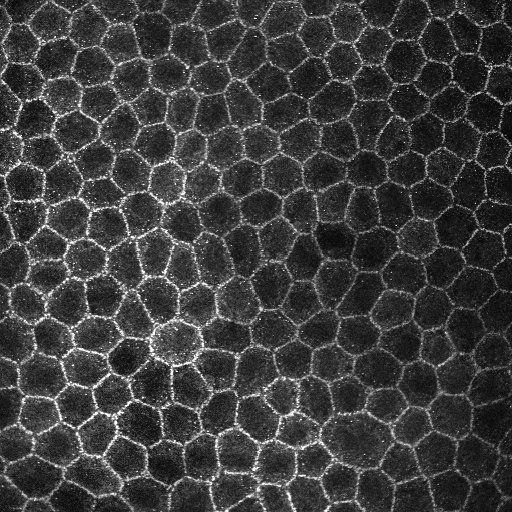 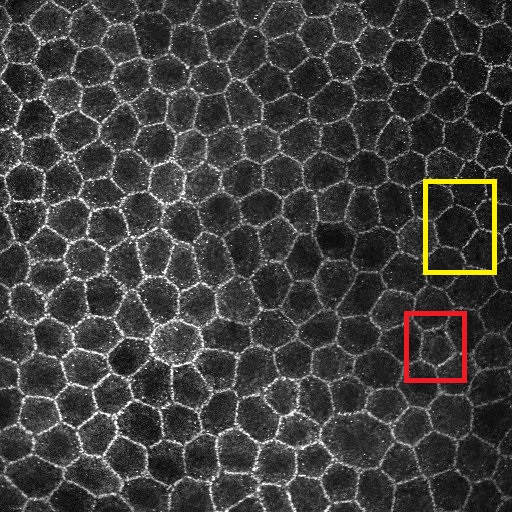 Any help is appreciated! NB I have looked at Gary Chinga's shape descriptor plugin and its not what I'm looking for =) |
|
One thing that we do is to :
a) find the cell centers (usually manually, or at best semi-automatically) b) compute a Voronoi Diagram (this usually looks VERY MUCH, but NOT IDENTICAL) to the cell boundaries c) histogram the #of neighbors for each cell center (that is, the number of sides to the Voronoi Cell associated with that “site” d) analyze such numbers as the % of cells with 6 neighbors. This is always custom, and how you do each step depends on the images you have. -- Kenneth Sloan [hidden email] Vision is the art of seeing what is invisible to others. > On Jul 6, 2015, at 17:14 , Mr_Sonky <[hidden email]> wrote: > > Hi all, > > I'm looking at some endothelial cells which are meant to be hexagonal and > tightly packed in nature. With stress and/or trauma, these cells exhbit > polymorphism - they turn into polygons of varying types. My aim is to > determine how many cells have lost their hexagon shapes in order to infere > the severity of the insult. I've attached two samples. The first is simply > the cellls. The second has a yellow box to highlight a Hexagon whilst the > red box highlights a cell that has undergone transformation into a a > different polygon (pentagon - in this instance). > > <http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell.jpg> > > <http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell_example.jpg> > > Any help is appreciated! > > NB I have looked at Gary Chinga's shape descriptor plugin and its not what > I'm looking for =) > > > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Index-of-hexagonality-tp5013442.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Mr_Sonky
It amounts to counting the number of neighbors each cell has.
for the images you provide it looks fairly simple to identify the cell outlines - edit and threshold this to produce a binary image showing the outlines thin to an 8 connected single line - (process binary skeletonize - set binary options to skeletonize with a dark background) should do it identify the nodes, where three cells meet (process binary option erode count 6 black background), can be easily identified from the number of connections between pixels on the thinned binarized image - at each node there will be a pixel with 3 or 4 neighbors. After the erosion each node will be a single pixel or group of pixels You now need the number of nodes per cell - threshold the binarized image to show the cells rather than the membrane, use (analyze particles) to produce a ROI for each cell - (exclude cells in the edge) by selecting the option to add each ROI to the ROI maanager Run through all the ROIs in the ROI manager and use the ROI for each cell (analyze particles) (again) to count the number of nodes - to do this either swell the ROI (edit selection enlarge) by a few pixels so that the nodes fall within the ROI, or swell (process binary dilate) the image of the nodes so that they overlap the ROI of the cells. ________________________________________ From: ImageJ Interest Group [[hidden email]] on behalf of Mr_Sonky [[hidden email]] Sent: 07 July 2015 00:14 To: [hidden email] Subject: Index of hexagonality? Hi all, I'm looking at some endothelial cells which are meant to be hexagonal and tightly packed in nature. With stress and/or trauma, these cells exhbit polymorphism - they turn into polygons of varying types. My aim is to determine how many cells have lost their hexagon shapes in order to infere the severity of the insult. I've attached two samples. The first is simply the cellls. The second has a yellow box to highlight a Hexagon whilst the red box highlights a cell that has undergone transformation into a a different polygon (pentagon - in this instance). <http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell.jpg> <http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell_example.jpg> Any help is appreciated! NB I have looked at Gary Chinga's shape descriptor plugin and its not what I'm looking for =) -- View this message in context: http://imagej.1557.x6.nabble.com/Index-of-hexagonality-tp5013442.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Mr_Sonky
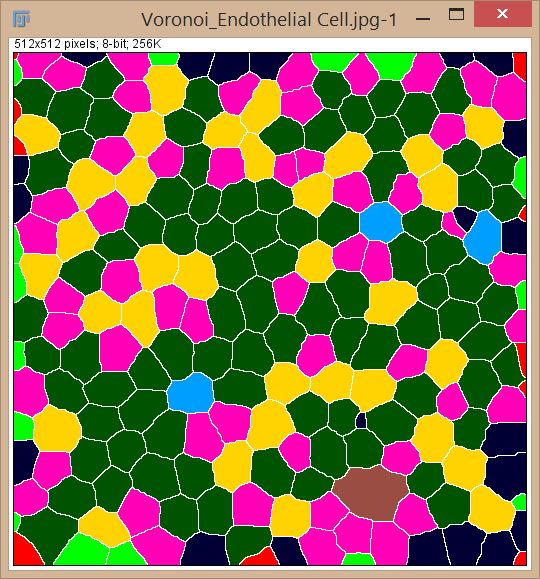
Hi Mr_Sonky,
The attatched Macro prepares the image you sent for the final analysis. This, might need adjustment depending on how all your other images look like. To run the macro you need the "BioVoxxel Toolbox" ( http://www.biovoxxel.de/development/) since it depends on veveral of its functions. At the end you can then run the "Neighbor Analysis" macro from the very same toolbox. Attatched you also find an image which more or less shows you the procedure and the outcome. Hope it helps, Jan //-----macro start original = getTitle(); run("8-bit"); run("Pseudo flat field correction", "blurring=40"); close(original + "_background"); selectWindow(original); run("Median...", "radius=1"); run("Select None"); run("Duplicate...", "title=Background"); background = getTitle(); run("Median...", "radius=40"); imageCalculator("Subtract", original, background); selectWindow(original); run("Enhance Contrast...", "saturated=1 normalize"); run("Auto Threshold", "method=Li white"); run("EDM Binary Operations", "iterations=2 operation=close"); run("Analyze Particles...", "size=50-Infinity show=Masks in_situ"); run("Median...", "radius=5"); run("Invert"); run("Watershed"); run("Voronoi"); setOption("BlackBackground", true); setThreshold(1, 255); run("Convert to Mask"); resetThreshold(); run("Invert"); close(background); selectWindow(original); //-----macro end 2015-07-07 0:14 GMT+02:00 Mr_Sonky <[hidden email]>: > Hi all, > > I'm looking at some endothelial cells which are meant to be hexagonal and > tightly packed in nature. With stress and/or trauma, these cells exhbit > polymorphism - they turn into polygons of varying types. My aim is to > determine how many cells have lost their hexagon shapes in order to infere > the severity of the insult. I've attached two samples. The first is simply > the cellls. The second has a yellow box to highlight a Hexagon whilst the > red box highlights a cell that has undergone transformation into a a > different polygon (pentagon - in this instance). > > <http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell.jpg> > > < > http://imagej.1557.x6.nabble.com/file/n5013442/Endothelial_Cell_example.jpg > > > > Any help is appreciated! > > NB I have looked at Gary Chinga's shape descriptor plugin and its not what > I'm looking for =) > > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Index-of-hexagonality-tp5013442.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
This post was updated on .
Hi all,
Firstly, thank you to those who have responded. Kenneth - Ive got to the Voronoi step but failed at the get neigbours bit. I think that requires the Biovoxxel plugin as Jan have provided below. Jeremy, thanks for your help too - it makes a lot of sense mathematically but I cant see a way to isolate the nodes. I'm probably too new at FIJI. Jan, I've run your script and it worked perfectly  . I got the exact same results as you have shown in your attachment. . I got the exact same results as you have shown in your attachment.
 Thank you all for your help! |
|
In reply to this post by gankaku
Thank you so much! I had the same problem and luckily I found this thread.. Thanks Jan!!
|
|
In reply to this post by gankaku
Hi,
I'm glad that I found this post. I'm trying to do exactly the same type of analysis so I installed the toolbox and the macro you provided. But somehow I still couldn't get the results right (for example I always get many cells with >40 edges/neighbors). Could you please let me know the steps to process the images to reach the correct measurements? I would start with a regular RGB image opened in Fiji, adjust threshold, run the macro to outline hexagon ect, and then run the "neighbor analysis" in the toolbox. I choose "Voronoi" as the type of analysis. It doesn't seem to matter how much I enter for "neighborhood radius" and the results are always screwed. Any advice would be much appreciated! Cheers, Hippo |
|
In reply to this post by gankaku
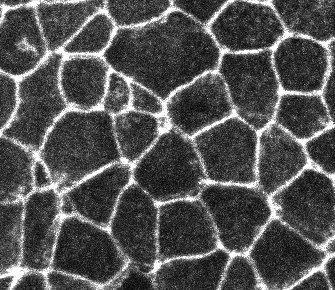
I just wanted to post a couple of examples of images and results I got. Let me know if you think any of the steps were not correct.
 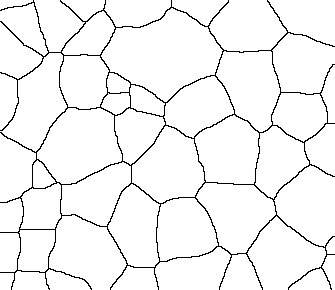 
|
Re: Index of hexagonality? - SOLVED
|
In reply to this post by Mr_Sonky
Hi,
I'm glad that I found this post. I'm trying to do exactly the same type of analysis so I installed the toolbox and the macro you provided. But somehow I still couldn't get the results right (for example I always get many cells with >40 edges/neighbors). Could you please let me know the steps to process the images to reach the correct measurements? I would start with a regular RGB image opened in Fiji, adjust threshold, run the macro to outline hexagon ect, and then run the "neighbor analysis" in the toolbox. I choose "Voronoi" as the type of analysis. It doesn't seem to matter how much I enter for "neighborhood radius" and the results are always screwed. Any advice would be much appreciated! Cheers, Hippo |
|
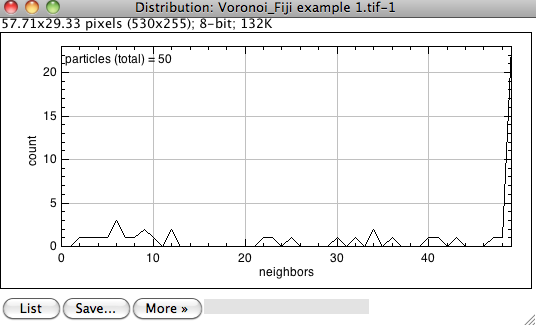
Hi hippo,
for the binary image you posted I get the result as expected It might be that the macro I provided for Mr_Sonky is not appropriate for your type of images and results in many very small or tiny particles which then obviously are included in the neighbor analysis. If you could provide an image of yours I could have a look again. @Mr_Sonky: Actually, since you get the distribution plot you can just click on the list button, transfer the list to excel and easily calculate the respective ratio od percentage for each amount of neighbors you have in respect to the overall number. Cheers, Jan 2016-03-29 18:12 GMT+02:00 cilia huang <[hidden email]>: > Hi, > > I'm glad that I found this post. I'm trying to do exactly the same type of > analysis so I installed the toolbox and the macro you provided. But somehow > I still couldn't get the results right (for example I always get many cells > with >40 edges/neighbors). Could you please let me know the steps to > process > the images to reach the correct measurements? I would start with a regular > RGB image opened in Fiji, adjust threshold, run the macro to outline > hexagon > ect, and then run the "neighbor analysis" in the toolbox. I choose > "Voronoi" > as the type of analysis. It doesn't seem to matter how much I enter for > "neighborhood radius" and the results are always screwed. Any advice would > be much appreciated! > > Cheers, > Hippo > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Index-of-hexagonality-SOLVED-tp5013442p5016023.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Index of hexagonality? - SOLVED
|
Hi Jan,
Thanks very much for getting back to me. I noticed that the unites of length for my images were in micron, and when I changed it to pixel I was able to get the proper results. I'm hoping if you could help me figure out another technical issue with the macro. I have some cells that have vertexes with 4 or more edges (see attached images) and irregular shape, which is part of the phenotype I'm trying to analyze. I feel like the macro is having difficulties picking 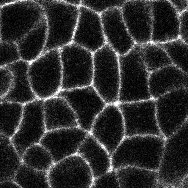 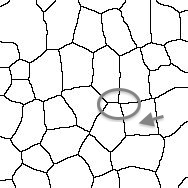 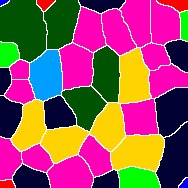 up those vertexes and gives me somewhat arbitrary results. For my control samples most cells are hexagon so there is no problem. up those vertexes and gives me somewhat arbitrary results. For my control samples most cells are hexagon so there is no problem.
In the images the circle marks the vertex, and the arrow points to a cell that I think has 4 neighbors...Let me know what you think. Thanks very much |
Re: Index of hexagonality? - SOLVED
|
On Wednesday 30 Mar 2016 16:17:13 you wrote:
> Hi Jan, > > Thanks very much for getting back to me. I noticed that the unites of length > for my images were in micron, and when I changed it to pixel I was able to > get the proper results. > > I'm hoping if you could help me figure out another technical issue with the > macro. I have some cells that have vertexes with 4 or more edges (see > attached images) and irregular shape, which is part of the phenotype I'm > trying to analyze. I feel like the macro is having difficulties picking > <http://imagej.1557.x6.nabble.com/file/n5016032/raw.jpg> > <http://imagej.1557.x6.nabble.com/file/n5016032/mask.jpg> > <http://imagej.1557.x6.nabble.com/file/n5016032/color_code_pattern.jpg> up > those vertexes and gives me somewhat arbitrary results. For my control > samples most cells are hexagon so there is no problem. > > In the images the circle marks the vertex, and the arrow points to a cell > that I think has 4 neighbors...Let me know what you think. than 4 neigbours. You could run a closing (a minimum filter of radius size (let's say) 5, followed by a maximum filter of the same size). That should round the corners of the tiles and leave the larger boundaries (see attached, unfortunately that has some extra pixels due to the grey circle and arrow in the original). The larger the radius of the opening, the more it will round the corners. Cheers Gabriel -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
On top of Gabriel's suggestion which you can easily achieve running the
"EDM Binary Operations" from the toolbox wit a iteration number of around 4-5 you may need to run the "Neighbor Analysis" with the setting "particle neighborhood" instead of "voronoi". Voronoi would mostly ignore the rounded corners but if you use the particle neighborhood with a pixel size of 2 then it should work. regards, Jan 2016-03-31 9:16 GMT+02:00 Gabriel Landini <[hidden email]>: > On Wednesday 30 Mar 2016 16:17:13 you wrote: > > Hi Jan, > > > > Thanks very much for getting back to me. I noticed that the unites of > length > > for my images were in micron, and when I changed it to pixel I was able > to > > get the proper results. > > > > I'm hoping if you could help me figure out another technical issue with > the > > macro. I have some cells that have vertexes with 4 or more edges (see > > attached images) and irregular shape, which is part of the phenotype I'm > > trying to analyze. I feel like the macro is having difficulties picking > > <http://imagej.1557.x6.nabble.com/file/n5016032/raw.jpg> > > <http://imagej.1557.x6.nabble.com/file/n5016032/mask.jpg> > > <http://imagej.1557.x6.nabble.com/file/n5016032/color_code_pattern.jpg> > up > > those vertexes and gives me somewhat arbitrary results. For my control > > samples most cells are hexagon so there is no problem. > > > > In the images the circle marks the vertex, and the arrow points to a cell > > that I think has 4 neighbors...Let me know what you think. > > In that version of the discrete space tessellation, the cell has indeed > more > than 4 neigbours. > You could run a closing (a minimum filter of radius size (let's say) 5, > followed by a maximum filter of the same size). That should round the > corners > of the tiles and leave the larger boundaries (see attached, unfortunately > that > has some extra pixels due to the grey circle and arrow in the original). > The > larger the radius of the opening, the more it will round the corners. > > Cheers > > Gabriel > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Index of hexagonality? - SOLVED
|
Awesome! Thank you so much! I'll power through the data set and let you know if there is anything else.
Cheers, Hippo |
Re: Index of hexagonality? - SOLVED
|
In reply to this post by Gabriel Landini
That's great;everything seems to be working now. Thank you!
|
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

