Modify (or create) results table
Modify (or create) results table
|
Hello,
I'm a new ImageJ user and complete newb when it comes macros etc. I use ImageJ to determine density, size of cell clusters in tissue sections. The macro I'm using (provided to me) is: macro "mac agg" {dir = getDirectory("Choose Source Directory"); list = getFileList(dir); for (i=0; i<list.length; i++) { open(dir+list[i]); run("8-bit"); setAutoThreshold("Default"); run("Threshold..."); waitForUser("Adjust threshold, then click 'OK' "); run("Analyze Particles...", "size=50-Infinity display exclude include summarize");close ();} saveAs("Measurements", "C:\\Documents and Settings\\mmatsche\\Desktop\\test.xls"); close();close();close(); }; This lets me pick a folder of images and do analyze particles in each image contained within. Results are added to the results table and a summary table is built as well. What I would like is to modify the results table (or create a new one if that is easier) but after looking through several examples online I couldn't figure it out. The first image posted here is an example of the results that I currently get (default results table). Unfortunately I no longer have a colleague with the skills to write macros. <nabble_img src="results.png" border="0"/> The "Label" is the image filename that the area of cell cluster is measured from. I can get anywhere from 20-200 cell clusters per image. I would like to change the "Label" column name to "Fish" and modify how ImageJ records the file name. Essentially, I would like to truncate the file name from (example) 14ST238-1.tif to 14ST238 (remove the dash and everything after). I don't need the file extension and the "-1" just before the .tif in the file name indicates the section number (I use 3 different images, or sections, per fish). Second, I would like to create a new column after the "Label" named "section". The "-1" just before the .tif extension is the section number. Can that number be identified and moved to the new column called "section"? Example. If I have 20 cell clusters in image 14ST150-2.tif, I would like all 20 measures of area and perim to be labeled as 14ST150 in the "Fish" column and "2" in the section column. These modifications would greatly simplify handling the output. Thanks! Mark M |
Re: Modify (or create) results table
|
Hi Mark,
I will try to answer but the picture of your result table is not displayed on the website. Do you only use the summary of results per image or do you need to have the complete detail of clusters? I don't think you can edit the "Label" heading of the column but you can modify the text content: if (nResults != 0){ for (i=0; i<nResults; i++){ resLabel = getResultLabel(i); if (indexOf(resLabel, ".tif") > 0) { resLabel = substring (resLabel, 0, lastIndexOf(resLabel,".tif")); // remove the ".tif" in the text string n = lengthOf(resLabel); // number of characters in the text string LBL = substring (resLabel, 0, n-2); // remove the last 2 characters to get the label section = substring (resLabel, n-1, n); // remove all text in label except last character setResult("Label", i, LBL); // set the text string for new label setResult("Section", i, section); // set the text string for section } } else { showMessage("No Result found in result table"); } This code block will search for ".tif" in each line of result label and truncate it. Then keep the last digit as the section and update each line with label and section. You can use: setResult("Label", i, "Fish"+LBL); to display the fish information. I hope this is what you needed. Cyril |
Re: Modify (or create) results table
|
Hi Cyril, Thanks so much for your help! The image was simply the default results table that is generated. I use both summary data (number of clusters per image and % are) and I use the areas of individual clusters. This is where I inserted the text you provided. Is this the correct place in the macro? I ran it and it gave an error "Else without if in line 82". macro "mac agg" {dir = getDirectory("Choose Source Directory"); list = getFileList(dir); for (i=0; i<list.length; i++) { open(dir+list[i]); run("8-bit"); setAutoThreshold("Default"); run("Threshold..."); waitForUser("Adjust threshold, then click 'OK' "); run("Analyze Particles...", "size=50-Infinity display exclude include summarize");close ();} if (nResults != 0){ for (i=0; i<nResults; i++){ resLabel = getResultLabel(i); if (indexOf(resLabel, ".tif") > 0) { resLabel = substring (resLabel, 0, lastIndexOf(resLabel,".tif")); // remove the ".tif" in the text string n = lengthOf(resLabel); // number of characters in the text string LBL = substring (resLabel, 0, n-2); // remove the last 2 characters to get the label section = substring (resLabel, n-1, n); // remove all text in label except last character setResult("Label", i, LBL); // set the text string for new label setResult("Section", i, section); // set the text string for section } } else { showMessage("No Result found in result table"); } selectWindow("Results"); saveAs("Measurements", "C:\\Documents and Settings\\mmatsche\\Desktop\\test.xls"); close();close();close(); }; Mark Matsche, MS Fish and Wildlife Health Maryland Dept. Natural Resources Cooperative Oxford Lab 904 S. Morris Street Oxford, MD 21654 410-226-5421 x129 (Ph) 410-226-0120 (Fax) On Fri, Feb 6, 2015 at 11:25 AM, Cyril Turiès [via ImageJ] <[hidden email]> wrote: Hi Mark, |
Re: Modify (or create) results table
|
In reply to this post by Cyril Turiès
Success! It was just a matter of a missing bracket. Yes, this is just what I needed. Thank you very much! Mark Matsche, MS Fish and Wildlife Health Maryland Dept. Natural Resources Cooperative Oxford Lab 904 S. Morris Street Oxford, MD 21654 410-226-5421 x129 (Ph) 410-226-0120 (Fax) On Fri, Feb 6, 2015 at 1:09 PM, Mark Matsche -DNR- <[hidden email]> wrote:
|
Re: Modify (or create) results table
|
Hi Mark,
I'm glad this solution matches your needs, little help between fish scientists. To check brackets you can use a text editor like jEdit to write your macros. Regards, Cyril |
Re: Modify (or create) results table
|
Hello Cyril, I hope this isn't too much of an imposition but I have one more addition to my macro that would reduce the number of steps in analysis for some tasks. The summary already provides a mean for the size of the macrophage aggregates "average size" within the field of view. Can I add a column to the summary results to also provide for the standard deviation around the means? Thanks! Mark Matsche, MS Fish and Wildlife Health Maryland Dept. Natural Resources Cooperative Oxford Lab 904 S. Morris Street Oxford, MD 21654 410-226-5421 x129 (Ph) 410-226-0120 (Fax) On Mon, Feb 9, 2015 at 3:47 AM, Cyril Turiès [via ImageJ] <[hidden email]> wrote: Hi Mark, |
Re: Modify (or create) results table
|
Hello Mark,
I think this is possible but you will have to build your own summary table. There is no ready-to-use solution this time as it depends on your image analysis steps and what you have in your Results table. Here is a good example of macro and functions to build custom summary stat http://rsb.info.nih.gov/ij/macros/CustomTabStatFromResults.txt In the middle of this document there is a section: // --------------- tab functions ---------------// you can add the standard deviation formula you need in the "makeStatFromResults" function and keep it in a variable that you add in the TabWindow function. One another solution (the easier one) is to get the result calculated with the command: run("Summarize"); But if you need it in your summary table you still have to make a custom one. First problem if you analyse many images you have a lot of results from different images and you cannot run a standard deviation calculation on the whole results. Maybe there is a way to get this value from statistics but I don't know how. Cyril |
Re: Modify (or create) results table
|
Thanks for the information! Mark Mark Matsche, MS Fish and Wildlife Health Maryland Dept. Natural Resources Cooperative Oxford Lab 904 S. Morris Street Oxford, MD 21654 410-226-5421 x129 (Ph) 410-226-0120 (Fax) On Tue, Mar 10, 2015 at 8:56 AM, Cyril Turiès [via ImageJ] <[hidden email]> wrote: Hello Mark, |
|
In reply to this post by Mark Matsche
I have more or less the same problem. I am trying to modify the custom results table:http://rsb.info.nih.gov/ij/macros/CustomTabStatFromResults.txt so that I can create a customized results table with variables from a series of automated steps with individual results tables each. the problem I am having is that, only variables for the first table are included in the final customResultsTable. Here is an example of the first two steps:
dir1 = getDirectory("Choose Source Directory "); dir2 = getDirectory("Choose Destination Directory "); list = getFileList(dir1); setBatchMode(true); for (i=0; i<list.length; i++) { showProgress(i+1, list.length); open(dir1+list[i]); run("Min...", "value=140"); run("8-bit"); run("Auto Local Threshold", "method=Median radius=15 parameter_1=0 parameter_2=0 white"); setOption("BlackBackground", false); run("Set Scale...", "distance=1617 known=21.8 pixel=1 unit=cm"); run("Set Measurements...", "area perimeter shape feret's display redirect=None decimal=2"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.2-1.00 show=[Overlay Outlines] display include summary record in_situ"); makeStatFromResults (); if (i==0) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); run("Gaussian Blur...", "sigma=50"); run("Make Binary"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.20-1.00 show=[Overlay Outlines] display include in_situ"); makeStatFromResults (); if (i==0) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); |
Re: Modify (or create) results table
|
Hi, I tried your macro and made some adjustments:
The "Auto Local Threshold" didn't work so I replaced it with "setAutoThreshold". You may have to check if it corresponds to your needs. You will find modifications in //comments// that you can delete or edit. You have access to all CustomTabStatFromResults functions as I copied/pasted it below "tab functions" from Gilles Carpentier's macro. Tell me if it works for you. ____________________________________________________________________ // tab variables var windowTabName="Stat Results Table",nameOfStatTab="["+windowTabName+"]",label="",undoErease=""; // stat variables var nbPerim=0,TheTotalArea=0,meanPerim=0,meanObject=0; dir1 = getDirectory("Choose Source Directory "); // Do you use it after?//dir2 = getDirectory("Choose Destination Directory "); list = getFileList(dir1); setBatchMode(true); for (i=1; i<list.length; i++) { showProgress(i+1, list.length); open(dir1+list[i]); run("Min...", "value=140"); run("8-bit"); // Changed to AutoThreshold// run("Auto Local Threshold", "method=Median radius=15 parameter_1=0 parameter_2=0 white"); setAutoThreshold("Mean"); setOption("BlackBackground", false); run("Set Scale...", "distance=1617 known=21.8 pixel=1 unit=cm"); run("Set Measurements...", "area perimeter shape feret's display redirect=None decimal=2"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.2-1.00 show=[Overlay Outlines] display include summary record in_situ"); makeStatFromResults (); //removed //if (i==1) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); run("Gaussian Blur...", "sigma=50"); run("Make Binary"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.20-1.00 show=[Overlay Outlines] display include in_situ"); makeStatFromResults (); //removed //if (i==1) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); } // --------------- tab functions ---------------// //CustomTabStatFromResults // Author : Gilles Carpentier // Faculte des Sciences et Technologies, // Universite Paris 12 Val de Marne, France. // function making stats from ImageJ Results Table values function makeStatFromResults () { sumObject=0;sumPerim=0;TheTotalArea=0;meanObject=0;meanPerim=0; // extraction from data from the Result Table: if (nResults() > 0 && isOpen("Results")) { if (columnLabelList ("Label") >=0) {label=getResultLabel (0);} else {label="unknown";} if (columnLabelList ("Area") >= 0) {sumObject=sumColumnTab ("Area"); } else {exit ("No \"Area\" measurements in this Result Table");} if (columnLabelList ("Perim.") >= 0) {sumPerim=sumColumnTab ("Perim."); } else {exit ("No \"Perim\.\" measurements in this Result Table");} } else {exit ("No result table");} // Stat calculations: TheTotalArea = sumObject; if (nResults() != 0) meanObject = TheTotalArea/nResults(); if (nResults() != 0) meanPerim=sumPerim/nResults(); } // function getting the sum of the value from the column name contained in kind function sumColumnTab (kind) { sum=0; if (columnLabelList (kind) >=0) { for (a=0; a<nResults(); a++) { sum=sum+getResult(kind,a); } return sum; } // function returning the number of the column which name is contained in kind. return -1 if doesn't exists function columnLabelList (kind) { columnNumber=-1; if (nResults() > 0 && isOpen("Results")) { selectWindow("Results"); results = getInfo(); lines = split(results, "\n"); headings = lines[0]; titlesofcolumns = split(headings, ",\t"); for (a=0; a<titlesofcolumns.length; a++) {if (titlesofcolumns[a] == kind) columnNumber=a;} } return columnNumber; } // function building/managing a table window function TabWindow (addLine) { undoErease = ""; if (! isOpen(windowTabName)) { run("New... ", "name="+nameOfStatTab+" type=Table"); print(nameOfStatTab, "\\Headings:Slice Name\tCount Objects\tTotal Area\tAverage Size\tMean Perim."); } if (addLine == 0) {print(nameOfStatTab, "\\Clear");} print(nameOfStatTab, label+ "\t" + nResults() + "\t" + TheTotalArea + "\t" + d2s(meanObject,2) + "\t" + d2s(meanPerim,2)); } // function removing the last line of the tab function rmLastLine () { if ( isOpen (windowTabName)) { selectWindow (windowTabName); tabContent = getInfo(); linesInTab = split(tabContent, "\n"); if (linesInTab[linesInTab.length-1] > 0) { print(nameOfStatTab, "\\Clear"); resteLines=""; for (i=1; i < (linesInTab.length -1); i++) { resteLines=resteLines+linesInTab[i] +"\n"; } if (linesInTab.length > 2) print (nameOfStatTab,resteLines); if (linesInTab.length > 1) undoErease=linesInTab[linesInTab.length-1]; } } } // function restoring the last ereased line in the table function undormLastLine () { if (undoErease != "") print(nameOfStatTab,undoErease); undoErease=""; } // saving a tab as excel type file function saveTab (path) { if (isOpen(windowTabName)) { if (path == "") { selectWindow (windowTabName); run("Input/Output...", "jpeg=75 gif=-1 file=.xls"); saveAs("Text"); } if (path != "") { selectWindow(windowTabName); saveAs("Text", path+windowTabName+".xls"); } } } function openTab (path,name) { undoErease="";windowTabName=name; lines=split(File.openAsString(path), "\n"); if (lines.length < 2) { exit ("This file doesn't seam to contain data");} headings = lines[0]; titlesOfColumns = split(headings, ",\t"); nameOfStatTab="["+windowTabName+"]"; if (isOpen(windowTabName)) {selectWindow(windowTabName) ;run("Close");} firstLine=""; for (i=0; i < (titlesOfColumns.length ); i++) { firstLine=firstLine+ titlesOfColumns [i]; if ( i < (titlesOfColumns.length )-1) {firstLine=firstLine+ "\t";} } toPrint=""; for (i=1; i < (lines.length ); i++) { toPrint=toPrint+lines[i]+"\n"; } run("New... ", "name="+nameOfStatTab+" type=Table"); print(nameOfStatTab, "\\Headings:"+firstLine+""); print(nameOfStatTab,toPrint); } |
|
Hie Cyril
Thank you for your assistance. I am trying to run the new code with your adjustments to see whether the variables from the two result tables can be presented in a new table. Unfortunately the code is not producing any output table. Did it work with you? Date: Tue, 13 Oct 2015 02:14:36 -0700 From: [hidden email] To: [hidden email] Subject: Re: Modify (or create) results table Hi, I tried your macro and made some adjustments: The "Auto Local Threshold" didn't work so I replaced it with "setAutoThreshold". You may have to check if it corresponds to your needs. You will find modifications in //comments// that you can delete or edit. You have access to all CustomTabStatFromResults functions as I copied/pasted it below "tab functions" from Gilles Carpentier's macro. Tell me if it works for you. ____________________________________________________________________ // tab variables var windowTabName="Stat Results Table",nameOfStatTab="["+windowTabName+"]",label="",undoErease=""; // stat variables var nbPerim=0,TheTotalArea=0,meanPerim=0,meanObject=0; dir1 = getDirectory("Choose Source Directory "); // Do you use it after?//dir2 = getDirectory("Choose Destination Directory "); list = getFileList(dir1); setBatchMode(true); for (i=1; i<list.length; i++) { showProgress(i+1, list.length); open(dir1+list[i]); run("Min...", "value=140"); run("8-bit"); // Changed to AutoThreshold// run("Auto Local Threshold", "method=Median radius=15 parameter_1=0 parameter_2=0 white"); setAutoThreshold("Mean"); setOption("BlackBackground", false); run("Set Scale...", "distance=1617 known=21.8 pixel=1 unit=cm"); run("Set Measurements...", "area perimeter shape feret's display redirect=None decimal=2"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.2-1.00 show=[Overlay Outlines] display include summary record in_situ"); makeStatFromResults (); //removed //if (i==1) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); run("Gaussian Blur...", "sigma=50"); run("Make Binary"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.20-1.00 show=[Overlay Outlines] display include in_situ"); makeStatFromResults (); //removed //if (i==1) TabWindow (0); else TabWindow (1); selectWindow("Results"); run("Close"); } // --------------- tab functions ---------------// //CustomTabStatFromResults // Author : Gilles Carpentier // Faculte des Sciences et Technologies, // Universite Paris 12 Val de Marne, France. // function making stats from ImageJ Results Table values function makeStatFromResults () { sumObject=0;sumPerim=0;TheTotalArea=0;meanObject=0;meanPerim=0; // extraction from data from the Result Table: if (nResults() > 0 && isOpen("Results")) { if (columnLabelList ("Label") >=0) {label=getResultLabel (0);} else {label="unknown";} if (columnLabelList ("Area") >= 0) {sumObject=sumColumnTab ("Area"); } else {exit ("No \"Area\" measurements in this Result Table");} if (columnLabelList ("Perim.") >= 0) {sumPerim=sumColumnTab ("Perim."); } else {exit ("No \"Perim\.\" measurements in this Result Table");} } else {exit ("No result table");} // Stat calculations: TheTotalArea = sumObject; if (nResults() != 0) meanObject = TheTotalArea/nResults(); if (nResults() != 0) meanPerim=sumPerim/nResults(); } // function getting the sum of the value from the column name contained in kind function sumColumnTab (kind) { sum=0; if (columnLabelList (kind) >=0) { for (a=0; a<nResults(); a++) { sum=sum+getResult(kind,a); } return sum; } // function returning the number of the column which name is contained in kind. return -1 if doesn't exists function columnLabelList (kind) { columnNumber=-1; if (nResults() > 0 && isOpen("Results")) { selectWindow("Results"); results = getInfo(); lines = split(results, "\n"); headings = lines[0]; titlesofcolumns = split(headings, ",\t"); for (a=0; a<titlesofcolumns.length; a++) {if (titlesofcolumns[a] == kind) columnNumber=a;} } return columnNumber; } // function building/managing a table window function TabWindow (addLine) { undoErease = ""; if (! isOpen(windowTabName)) { run("New... ", "name="+nameOfStatTab+" type=Table"); print(nameOfStatTab, "\\Headings:Slice Name\tCount Objects\tTotal Area\tAverage Size\tMean Perim."); } if (addLine == 0) {print(nameOfStatTab, "\\Clear");} print(nameOfStatTab, label+ "\t" + nResults() + "\t" + TheTotalArea + "\t" + d2s(meanObject,2) + "\t" + d2s(meanPerim,2)); } // function removing the last line of the tab function rmLastLine () { if ( isOpen (windowTabName)) { selectWindow (windowTabName); tabContent = getInfo(); linesInTab = split(tabContent, "\n"); if (linesInTab[linesInTab.length-1] > 0) { print(nameOfStatTab, "\\Clear"); resteLines=""; for (i=1; i < (linesInTab.length -1); i++) { resteLines=resteLines+linesInTab[i] +"\n"; } if (linesInTab.length > 2) print (nameOfStatTab,resteLines); if (linesInTab.length > 1) undoErease=linesInTab[linesInTab.length-1]; } } } // function restoring the last ereased line in the table function undormLastLine () { if (undoErease != "") print(nameOfStatTab,undoErease); undoErease=""; } // saving a tab as excel type file function saveTab (path) { if (isOpen(windowTabName)) { if (path == "") { selectWindow (windowTabName); run("Input/Output...", "jpeg=75 gif=-1 file=.xls"); saveAs("Text"); } if (path != "") { selectWindow(windowTabName); saveAs("Text", path+windowTabName+".xls"); } } } function openTab (path,name) { undoErease="";windowTabName=name; lines=split(File.openAsString(path), "\n"); if (lines.length < 2) { exit ("This file doesn't seam to contain data");} headings = lines[0]; titlesOfColumns = split(headings, ",\t"); nameOfStatTab="["+windowTabName+"]"; if (isOpen(windowTabName)) {selectWindow(windowTabName) ;run("Close");} firstLine=""; for (i=0; i < (titlesOfColumns.length ); i++) { firstLine=firstLine+ titlesOfColumns [i]; if ( i < (titlesOfColumns.length )-1) {firstLine=firstLine+ "\t";} } toPrint=""; for (i=1; i < (lines.length ); i++) { toPrint=toPrint+lines[i]+"\n"; } run("New... ", "name="+nameOfStatTab+" type=Table"); print(nameOfStatTab, "\\Headings:"+firstLine+""); print(nameOfStatTab,toPrint); } If you reply to this email, your message will be added to the discussion below:
http://imagej.1557.x6.nabble.com/Modify-or-create-results-table-tp5011481p5014615.html
|
RE: Modify (or create) results table
|
Hi, I tried this macro on 6 images and all the results were on the same stat table:
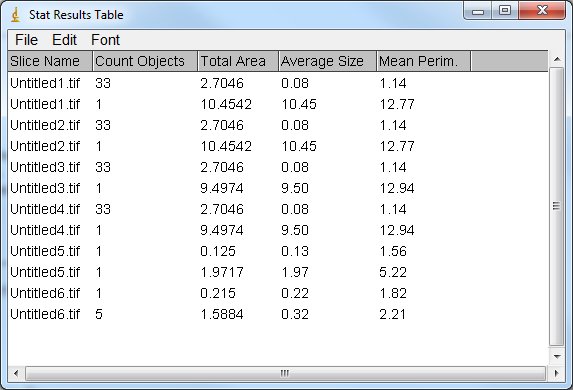 It was on ImageJ 1.50b Are you working on Fiji or ImageJ and on which version? Cyril |
|
Oh I see. Quite interesting. However I am using different variables for the second macro. So I would add the result in columns instead of rows. Is this possible?
Date: Thu, 15 Oct 2015 00:27:29 -0700 From: [hidden email] To: [hidden email] Subject: RE: Modify (or create) results table Hi, I tried this macro on 6 images and all the results were on the same stat table:  It was on ImageJ 1.50b Are you working on Fiji or ImageJ and on which version? Cyril If you reply to this email, your message will be added to the discussion below:
http://imagej.1557.x6.nabble.com/Modify-or-create-results-table-tp5011481p5014644.html
|
RE: Modify (or create) results table
|
If I understand your need it should be something like this:
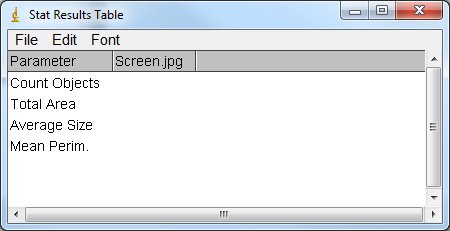 It's easy for the first column but if you add a new column for each image you analyze, it will be difficult to manage as you are not in "result table". In result table you can set result in a specific line or column "setResult("Column", row, value)", with custom tables it's only text strings. You will have to create the new heading and insert the data in the text string at the correct position. In your analysis, do you need the ImageJ result table or do you only need the custom stat table? |
|
Hie Cyril
I need the Stat Table and not the result table. With the Stat table I will be able to include some linear models to compute new variables from each macro. Date: Fri, 16 Oct 2015 06:59:59 -0700 From: [hidden email] To: [hidden email] Subject: RE: Modify (or create) results table If I understand your need it should be something like this:  It's easy for the first column but if you add a new column for each image you analyze, it will be difficult to manage as you are not in "result table". In result table you can set result in a specific line or column "setResult("Column", row, value)", with custom tables it's only text strings. You will have to create the new heading and insert the data in the text string at the correct position. In your analysis, do you need the ImageJ result table or do you only need the custom stat table? If you reply to this email, your message will be added to the discussion below:
http://imagej.1557.x6.nabble.com/Modify-or-create-results-table-tp5011481p5014662.html
|
RE: Modify (or create) results table
|
In reply to this post by rmakanza
Hi,
I tried a new version to edit the ImageJ Result table at the end of the macro. During the batch process results are added to arrays and after the last image the result table is updated with all measurements: 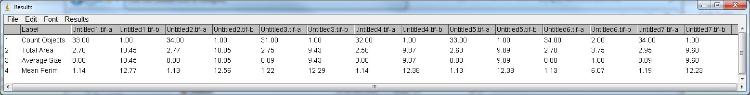 I did not test the macro on a directory containing sub-directories so be careful in that case. You have to paste the code from CustomTabStatFromResults below the "tab functions" line, I didn't copied it because it was unchanged. Just let me know if it works with your image analysis. Cyril ________________________________________ // tab variables var windowTabName="Stat Results Table",nameOfStatTab="["+windowTabName+"]",label="",undoErease=""; // stat variables var nbPerim=0,TheTotalArea=0,meanPerim=0,meanObject=0; dir1 = getDirectory("Choose Source Directory "); list = getFileList(dir1); setBatchMode(true); for (i=0; i<list.length; i++) { showProgress(i, list.length); open(dir1+list[i]); run("Min...", "value=140"); run("8-bit"); // Changed to AutoThreshold// run("Auto Local Threshold", "method=Median radius=15 parameter_1=0 parameter_2=0 white"); setAutoThreshold("Mean"); setOption("BlackBackground", false); run("Set Scale...", "distance=1617 known=21.8 pixel=1 unit=cm"); run("Set Measurements...", "area perimeter shape feret's display redirect=None decimal=2"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.2-1.00 show=[Overlay Outlines] display clear include record in_situ"); if (nResults() > 0 && isOpen("Results")) { makeStatFromResults (); TabWindow (1); } else { print(nameOfStatTab, label+"-No result");} // Arrays containing results of first particle analysis if (i == 0){ resLabel = newArray(""+label+"-a"); resNB = newArray(1); Array.fill(resNB, nResults()); resArea = newArray(1); Array.fill(resArea, TheTotalArea); resObject = newArray(1); Array.fill(resObject, d2s(meanObject,2)); resPerim = newArray(1); Array.fill(resPerim, d2s(meanPerim,2)); } else { resLabel = Array.concat(resLabel, label+"-a"); resNB = Array.concat(resNB, nResults()); resArea = Array.concat(resArea, TheTotalArea); resObject = Array.concat(resObject, d2s(meanObject,2)); resPerim = Array.concat(resPerim, d2s(meanPerim,2)); } run("Gaussian Blur...", "sigma=50"); run("Make Binary"); run("Analyze Particles...", "size=0.05-Infinity circularity=0.20-1.00 show=[Overlay Outlines] display clear include record in_situ"); if (nResults() > 0 && isOpen("Results")) { makeStatFromResults (); TabWindow (1); } else { print(nameOfStatTab, label+"-No result");} // Add results from second analysis to arrays resLabel = Array.concat(resLabel, label+"-b"); resNB = Array.concat(resNB, nResults()); resArea = Array.concat(resArea, TheTotalArea); resObject = Array.concat(resObject, d2s(meanObject,2)); resPerim = Array.concat(resPerim, d2s(meanPerim,2)); } // Add results from arrays to results table run("Clear Results"); run("Set Measurements...", " redirect=None decimal=2"); updateResults(); setResult("Label", 0, "Count Objects"); setResult("Label", 1, "Total Area"); setResult("Label", 2, "Average Size"); setResult("Label", 3, "Mean Perim"); updateResults(); for (a=0; a<resLabel.length; a++) { setResult(resLabel[a], 0, resNB[a]); setResult(resLabel[a], 1, resArea[a]); setResult(resLabel[a], 2, resObject[a]); setResult(resLabel[a], 3, resPerim[a]); updateResults(); } // --------------- tab functions ---------------// //CustomTabStatFromResults // Author : Gilles Carpentier // Faculte des Sciences et Technologies, // Universite Paris 12 Val de Marne, France. ..... insert here CustomTabStatFromResults functions |
RE: Modify (or create) results table
|
Sorry the screen is too small.
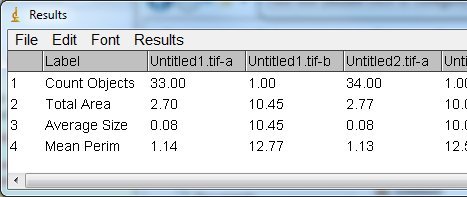 In addition, the macro should work on images with zero particle analysed, with a "-No result" text to the Stat Results Table. |
|
Hie Cyril
Thank you so much for your assistance. Let me try your new script. Best Regards Richard Date: Thu, 22 Oct 2015 02:16:07 -0700 From: [hidden email] To: [hidden email] Subject: RE: Modify (or create) results table Sorry the screen is too small.  In addition, the macro should work on images with zero particle analysed, with a "-No result" text to the Stat Results Table. If you reply to this email, your message will be added to the discussion below:
http://imagej.1557.x6.nabble.com/Modify-or-create-results-table-tp5011481p5014719.html
|
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

