Need help to analyze an image
|
Hello,
I'm new to imageJ and I need your help please !! I have attached my image and I want to separate this two cells, I found that analyze particle plugin can count the cells as i want but how i can separate them before using it? Thank you for any help or suggestions .. 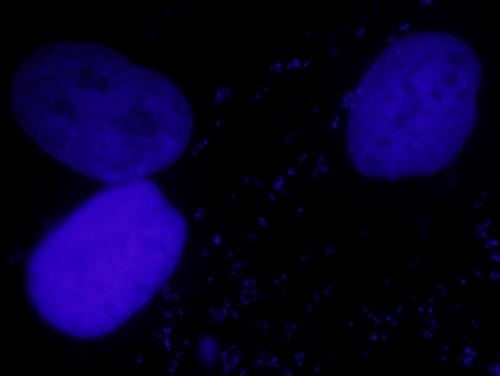
|
Re: Need help to analyze an image
|
Hi Nariman,
Probably the easiest is Process>Binary>Watershed on the thresholded image. Before this, it may help to run a slight background subtraction (Process>Subtract Background, sliding paraboloid, no light background, radius a few hundred). If you want to do measurements where the binary (thresholded) image is insufficient, use 'Redirect to' to the original data. Michael ________________________________________________________________ On 14 May 2010, at 12:16, Nariman wrote: > Hello, > > I'm new to imageJ and I need your help please !! > I have attached my image and I want to separate this two cells, I > found that > analyze particle plugin can count the cells as i want but how i can > separate > them before using it? > > Thank you for any help or suggestions .. > http://n2.nabble.com/file/n5050304/cells.jpg |
Re: Need help to analyze an image
|
Hi Nariman,
The attached macro will separate your nuclei. I recommend using a local threshold approach as you have a range of signals from the different nuclei. You'll have to download G. Landini's Auto_Local_Threshold .jar file, place it in the plugins folder of ImageJ then restart ImageJ before running the macro. To run the macro, select plugins>macros>run. Browse for the macro and select open. http://www.dentistry.bham.ac.uk/landinig/software/software.html Text of the attached macro: run("8-bit"); run("Auto Local Threshold", "method=MidGrey radius=5 parameter_1=0 parameter_2=0 white"); run("Analyze Particles...", "size=5000-Infinity circularity=0.00-1.00 show=Nothing clear summarize add"); run("Revert"); Best Wishes, Dan Hoeppner NIH On 5/14/10 8:43 AM, "Michael Schmid" <[hidden email]> wrote: Hi Nariman, Probably the easiest is Process>Binary>Watershed on the thresholded image. Before this, it may help to run a slight background subtraction (Process>Subtract Background, sliding paraboloid, no light background, radius a few hundred). If you want to do measurements where the binary (thresholded) image is insufficient, use 'Redirect to' to the original data. Michael ________________________________________________________________ On 14 May 2010, at 12:16, Nariman wrote: > Hello, > > I'm new to imageJ and I need your help please !! > I have attached my image and I want to separate this two cells, I > found that > analyze particle plugin can count the cells as i want but how i can > separate > them before using it? > > Thank you for any help or suggestions .. > http://n2.nabble.com/file/n5050304/cells.jpg |
|
Hi Daniel and Michael,
Indeed when I tried the same macro, I got 11 particles and they It doesn't encircle the nuclei !! (and when I applied it to my real image which is .tif I didn't get any result !!) Then I applied the same instruction that Michael told me about: run("Subtract Background...", "rolling=300 sliding"); Then I should apply Watershed on the thresholded image, I tried to threshold the image after applying "contrast enhance" and this didn't help me too. This is the macro that I apply it run("Enhance Contrast", "saturated=0.4"); run("Subtract Background...", "rolling=300 sliding"); run("MultiThresholder", "isodata"); run("8-bit"); run("Watershed"); Watershed didn't segment my cells well!! Is there any preprocessing could help me before that? Thank you so much for your help.. |
Re: Need help to analyze an image
|
In reply to this post by Nariman
On May 14, 2010, at 6:16 AM, Nariman wrote:
> Hello, > > I'm new to imageJ and I need your help please !! > I have attached my image and I want to separate this two cells, I found that > analyze particle plugin can count the cells as i want but how i can separate > them before using it? > > Thank you for any help or suggestions .. > http://n2.nabble.com/file/n5050304/cells.jpg You can separate the cells using Process>Binary>Watershed but some preprocessing (blurring) is required. Here is an example macro: rename("RGB"); // known name needed for redirect run("Duplicate...", "title=Mask"); run("8-bit"); run("Gaussian Blur...", "sigma=2"); setAutoThreshold("Li dark"); run("Convert to Mask"); run("Watershed"); run("Set Measurements...", "area mean min limit redirect=RGB"); run("Analyze Particles...", "size=1000-Infinity display clear"); -wayne |
|
Hello,
I'm sorry for asking again but I don't know why watershed separated the particles in this way (I'v attached my image) !! Maybe I should detect the edges before or something else? or there is another solution?? (I applied the same macro that Wayne wrote it, I added only "Subtract Background" before it and I tried to enhance the contrast but it was always the same results!!) Thank you so much for your help.. 
|
Re: Need help to analyze an image
|
I'm confused, as usual. Are you trying to do someting like partition the two
touching cells into two separate cells using this image? David On Sat, May 15, 2010 at 12:06 AM, Nariman <[hidden email]> wrote: > Hello, > I'm sorry for asking again but I don't know why watershed separated the > particles in this way (I'v attached my image) !! > Maybe I should detect the edges before or something else? or there is > another solution?? > (I applied the same macro that Wayne wrote it, I added only "Subtract > Background" before it and I tried to enhance the contrast but it was always > the same results!!) > Thank you so much for your help.. > http://n2.nabble.com/file/n5058431/Mask.png > -- > View this message in context: > http://imagej.588099.n2.nabble.com/Need-help-to-analyze-an-image-tp5050304p5058431.html > Sent from the ImageJ mailing list archive at Nabble.com. > |
Re: Need help to analyze an image
|
> I'm sorry for asking again but I don't know why watershed separated the
> particles in this way (I'v attached my image) !! > Maybe I should detect the edges before or something else? or there is > another solution?? > http://n2.nabble.com/file/n5058431/Mask.png It is hard to see in the png image (it seems to be downscaled) - there seems to be a segmentation at a constriction in one of the cells, so the standard Watershed command is too sensitive. See this post for a macro that gives you control over the sensitivity of watershed: https://list.nih.gov/cgi-bin/wa.exe?A2=ind0909&L=IMAGEJ&F=&S=&P=214248 Michael |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

