In all honesty your PI should be helping you with this and there are tutorials out there but hopefully this helps enough.
1) Open ImageJ
2) Open your desired image by dragging it over the ImageJ toolbar and dropping it
For the "Blue" Channel ONLY!3) Click Image -> Color -> Split channels
4) Click Image -> Adjust -> Threshold
5) Check "Dark Background" checkbox
6) Use the first slider bar to make sure that the cells nuclei are filled with red
7) Click "Apply"
8) Click Process -> Binary -> Watershed
9) Click Analyze -> Analyze Particles
10) Use the following settings
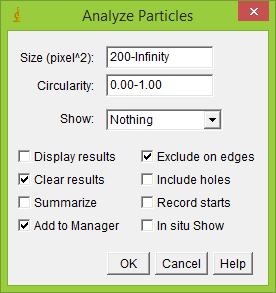
11) Click "Ok"
12) Analyze your nuclei, make sure there are no micro-nuclei or overlapping ones, or ones that encircle multiple nuclei. If there are click on them in the image and then click "delete" in the ROImanager.
For the desired channel with the "protein" in it only!13) You can then click on the image that has your proteins and click "Show all" in the ROImanager twice (once to click "off", another time to click "on" while you have the protein window selected!) and the overlay should apply to it.