Repeating ROI's in a cell
|
Hello everyone,
I am very new to ImageJ, but have some programming experience in python, and was seeking some advice. I am trying to quantify the change in intensity of a protein as a cell is migrating. I would like to create a macro similar to the one in the image. Basically, I would hand draw an ROI around a single cell. Then run a macro that would repeat a predefined size hexagon within the boundaries of my pre-drawn ROI. Then each ROI would record the mean intensities over 100-200 frames. Then I would group that data by intensity and duration of the intensity change. I am at a complete loss in how to do this. I could program in the hexagon ROI, but how do I loop that ROI around the predefined limits of a cell? Then, how do I get each ROI to record the intensity changes over time? Any help at all would be much appreciated! Thanks, Michelle 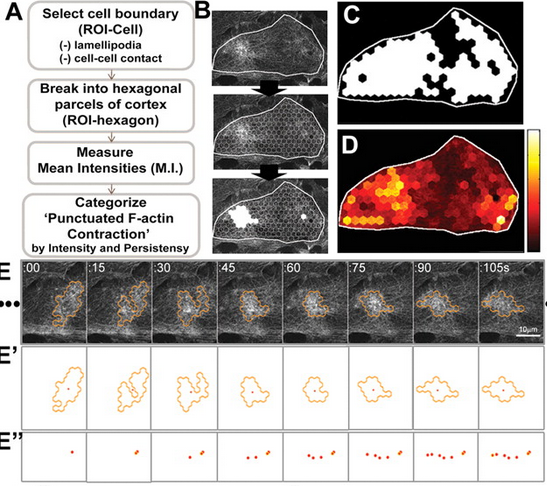
|
|
Hi Michelle,
> <http://imagej.1557.x6.nabble.com/file/n5010534/ImageJROI.png> linking to the original work [1] (and maybe even citing the authors' names: Kim and Davidson 2010), instead of just copying an image without asking, would be considered best scientific practice, and it would save time for people willing to help you. That said, did you contact the original authors about their method? According to the paper, they have used ImageJ, so they should even be able to provide you with their custom macros. Cheers, Jan [1]: http://jcs.biologists.org/content/124/4/635 On 18.11.14 23:34, MichelleL wrote: > Hello everyone, > > I am very new to ImageJ, but have some programming experience in python, and > was seeking some advice. I am trying to quantify the change in intensity of > a protein as a cell is migrating. I would like to create a macro similar to > the one in the image. Basically, I would hand draw an ROI around a single > cell. Then run a macro that would repeat a predefined size hexagon within > the boundaries of my pre-drawn ROI. Then each ROI would record the mean > intensities over 100-200 frames. Then I would group that data by intensity > and duration of the intensity change. > > I am at a complete loss in how to do this. I could program in the hexagon > ROI, but how do I loop that ROI around the predefined limits of a cell? > Then, how do I get each ROI to record the intensity changes over time? > > Any help at all would be much appreciated! > > Thanks, > > Michelle > > <http://imagej.1557.x6.nabble.com/file/n5010534/ImageJROI.png> > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Repeating-ROI-s-in-a-cell-tp5010534.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Jan,
Thank you for your response, and for posting the citation. I meant no offense, by not referencing the image, it was my mistake. I did contact the authors of the paper, and have not had a response. After spending some time trying to reproduce the code on my own, I thought I would ask for help from the community. Thanks! Michelle |
Re: Repeating ROI's in a cell
|
In reply to this post by MichelleL
Hi Michelle,
I have a few questions. >Basically, I would hand draw an ROI around a single cell. Will this be drawn for each time point? Or will you use the initial cell outline for the entire series? >Then run a macro that would repeat a predefined size hexagon within the boundaries of my pre-drawn ROI. Then each ROI would record the mean intensities over 100-200 frames. Will the ROIs need to move as the cell moves? This relates to my first question. >Then I would group that data by intensity and duration of the intensity change. Just to be clear. Group by intensity in each frame? Or total intensity over the course of the 100-200 frames? George On Tue, Nov 18, 2014 at 5:34 PM, MichelleL <[hidden email]> wrote: > Hello everyone, > > I am very new to ImageJ, but have some programming experience in python, > and > was seeking some advice. I am trying to quantify the change in intensity > of > a protein as a cell is migrating. I would like to create a macro similar > to > the one in the image. Basically, I would hand draw an ROI around a single > cell. Then run a macro that would repeat a predefined size hexagon within > the boundaries of my pre-drawn ROI. Then each ROI would record the mean > intensities over 100-200 frames. Then I would group that data by intensity > and duration of the intensity change. > > I am at a complete loss in how to do this. I could program in the hexagon > ROI, but how do I loop that ROI around the predefined limits of a cell? > Then, how do I get each ROI to record the intensity changes over time? > > Any help at all would be much appreciated! > > Thanks, > > Michelle > > <http://imagej.1557.x6.nabble.com/file/n5010534/ImageJROI.png> > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Repeating-ROI-s-in-a-cell-tp5010534.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Repeating ROI's in a cell
|
Hi Michelle,
This macro could get you started: https://gist.github.com/mutterer/035ade419bf9c96475ce I'm creating a mask with all hexagons, then use the traced selection and analyze particles with exclude flag to add relevant hexagons to the roi manager. From there, you have your hexagons in it, so you can reuse them to go and measure on whatever image(s). Sincerely, Jerome On 19 November 2014 16:03, George Patterson <[hidden email]> wrote: > Hi Michelle, > I have a few questions. > >Basically, I would hand draw an ROI around a single > cell. > Will this be drawn for each time point? Or will you use the initial cell > outline for the entire series? > > >Then run a macro that would repeat a predefined size hexagon within > the boundaries of my pre-drawn ROI. Then each ROI would record the mean > intensities over 100-200 frames. > Will the ROIs need to move as the cell moves? This relates to my first > question. > > >Then I would group that data by intensity > and duration of the intensity change. > Just to be clear. Group by intensity in each frame? Or total intensity > over the course of the 100-200 frames? > > George > > On Tue, Nov 18, 2014 at 5:34 PM, MichelleL <[hidden email]> wrote: > > > Hello everyone, > > > > I am very new to ImageJ, but have some programming experience in python, > > and > > was seeking some advice. I am trying to quantify the change in intensity > > of > > a protein as a cell is migrating. I would like to create a macro similar > > to > > the one in the image. Basically, I would hand draw an ROI around a > single > > cell. Then run a macro that would repeat a predefined size hexagon > within > > the boundaries of my pre-drawn ROI. Then each ROI would record the mean > > intensities over 100-200 frames. Then I would group that data by > intensity > > and duration of the intensity change. > > > > I am at a complete loss in how to do this. I could program in the > hexagon > > ROI, but how do I loop that ROI around the predefined limits of a cell? > > Then, how do I get each ROI to record the intensity changes over time? > > > > Any help at all would be much appreciated! > > > > Thanks, > > > > Michelle > > > > <http://imagej.1557.x6.nabble.com/file/n5010534/ImageJROI.png> > > > > > > > > -- > > View this message in context: > > > http://imagej.1557.x6.nabble.com/Repeating-ROI-s-in-a-cell-tp5010534.html > > Sent from the ImageJ mailing list archive at Nabble.com. > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- Jerome Mutterer CNRS - Institut de biologie moléculaire des plantes 12, rue du Général Zimmer 67084 Strasbourg Cedex T 0367155339 www.ibmp.cnrs.fr -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by George Patterson
Hi George,
Thank you for your help! I was planning on using the initial ROI for the entire time series. The cells are relatively non-motile and I am imaging for a short time interval, so a single ROI should be fine. The individual ROI's would not need to move as the cell moves, I just need a way to window the cell into predefined areas. My protein has a wave-like directionality, I would like to group the intensity in each frame, but also track the directionality of the intensity over time. I can do that by hand as well, as long as I can window the cell with a predefined size ROI. Michelle |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

