SKM Cross sectional area measurement
SKM Cross sectional area measurement
|
Hi,
I am trying to calculate the cross sectional area of skeletal muscle fibers obtained via muscle biopsy. I have tried to generate an automated macro but with little success. Given the large number of sections that I will have, and automated steps will make things a lot better and reduce error. Thanks for your advise. |
Re: SKM Cross sectional area measurement
|
On 29/01/2014 14:37, stevefaulkner wrote:
> Hi, > > I am trying to calculate the cross sectional area of skeletal muscle fibers > obtained via muscle biopsy. I have tried to generate an automated macro but > with little success. Given the large number of sections that I will have, > and automated steps will make things a lot better and reduce error. > > Thanks for your advise. > Hi Steve, Please could we see a sample image? I assume that you'd want to start by isolating the areas you wish to measure somehow. Usually, blurring then thresholding works well if the area of interest is clearly defined against the background. Once you know the steps to reliably separate the regions to measure, it should be simple to use analyse particles to measure the area. The macro recorder is very useful as a history while you're testing things out on some sample images. Once you've got the method worked out, you can copy and paste the relevant bits into a new macro. How are your results presented? Do you have a folder full of images or a stack? Your macro can process either, but the method differs. Hopefully you'll glean some useful info from a similar topic that prompted a post on our website: http://www.esimagingsolutions.com/resources/case-studies/119-case-study-labelling-a-moving-object-in-an-image-stack-with-imagej-macro Let us know if you're still stuck! Best, Ed -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: SKM Cross sectional area measurement
|
In reply to this post by stevefaulkner
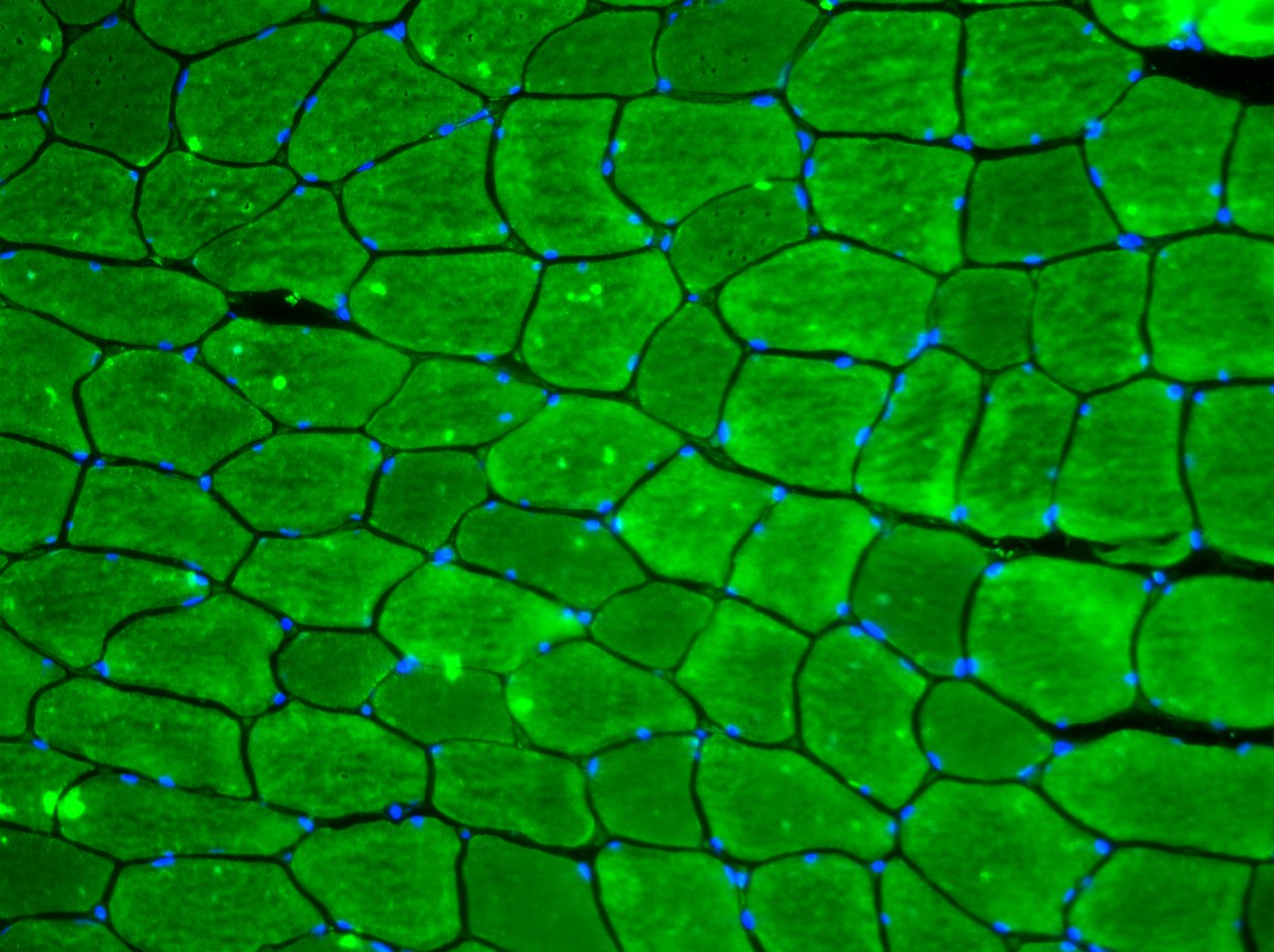 Ed, Thanks for your response. Any advice greatly appreciated. I'm trying to single out the muscle fibres and automatically generate an accurate measure of CSA that does not involve a manual (an possibly biased/unreliable) measurement. |
Re: SKM Cross sectional area measurement
|
In reply to this post by stevefaulkner
Yes they are all saved as tif files! The aim is to generate a CSA for each cell within the image, but ignoring those that come ito contact with the edges. So far I have been doing this by creating and ROI and then manually measuring a selections of cells, which is less than ideal! As you suggest, I have been doing this using the green channel alone, I just wanted to show a complete image as I'll also be analysing the density and activation of a number of other cell within the image.
When I have tried to threshold the image, the problem that I get is that the fluoro isn't uniform throughout, so each cell often becomes grainy and is no longer recogonised as a single entity as defined by the cell membrane. I will have a number of images to analyse but so far they are just stored as each composite image (each filter) of the total overlay in a single file from a single field of view, all within a subject specific folder. Steve |
Re: SKM Cross sectional area measurement
|
Hi Steve,
While I have not tried it myself, I think the trainable segmentation (weka) might be able to help: http://fiji.sc/Trainable_Weka_Segmentation I've had success with a similar, but non ImageJ product for trainable segementation as well called ilastik: http://ilastik.org/ On Thu, Jan 30, 2014 at 6:21 AM, stevefaulkner <[hidden email]>wrote: > Yes they are all saved as tif files! The aim is to generate a CSA for each > cell within the image, but ignoring those that come ito contact with the > edges. So far I have been doing this by creating and ROI and then manually > measuring a selections of cells, which is less than ideal! As you suggest, > I > have been doing this using the green channel alone, I just wanted to show a > complete image as I'll also be analysing the density and activation of a > number of other cell within the image. > > When I have tried to threshold the image, the problem that I get is that > the > fluoro isn't uniform throughout, so each cell often becomes grainy and is > no > longer recogonised as a single entity as defined by the cell membrane. > > I will have a number of images to analyse but so far they are just stored > as > each composite image (each filter) of the total overlay in a single file > from a single field of view, all within a subject specific folder. > > Steve > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/SKM-Cross-sectional-area-measurement-tp5006330p5006338.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

