Segmenting Circular Objects that are touching multiple other objects
|
Hello,
I am attempting to size microspheres that I have imaged on TEM. The microspheres are touching multiple other microspheres and I am having difficulty segmenting/thresholding the objects to maintain the original size of the microsphere. I either have the problem of the negative space becoming the main focus of the particle analysis, or I have thresholded the image so much that the particle size is no longer accurately measured. In other programs, there is the ability to automatically draw circles around an object based on the circularity of part of that that object. Is that ability available in ImageJ? Thank you. 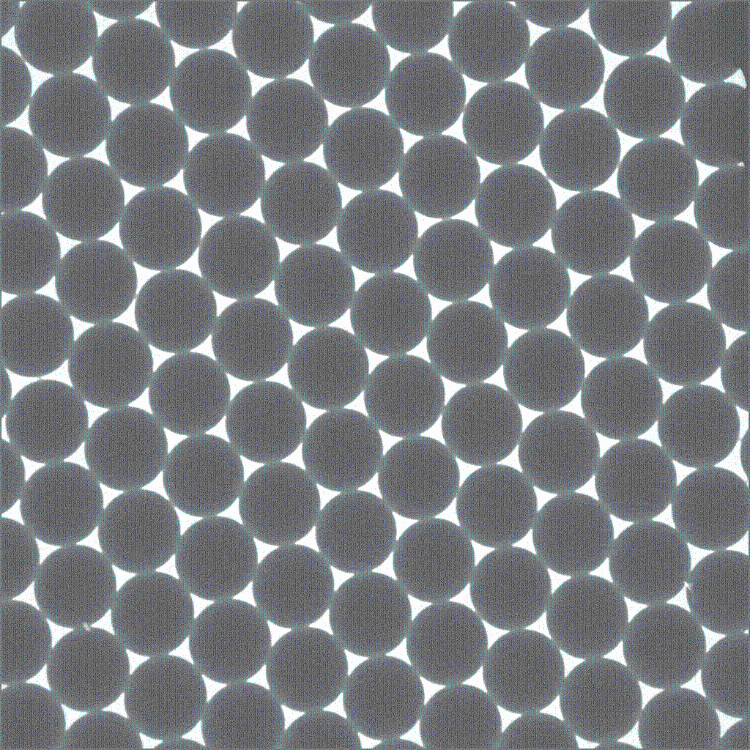
|
Re: Segmenting Circular Objects that are touching multiple other objects
|
Have you tried applying a watershed segmentation after your initial
thresholding to separate touching particles? I just tried it with your image after thresholding the image with the default settings and it appeared to do a good job. You can then use 'Analyse Particle' to add all your particles to the ROI manager and measure them. If you select 'Fit ellipse' in the measure option you will also get the length of the two main axes of the particle, while 'Centroid' or 'Centre of mass' provide the coordinates of the centre of each particle. Based on the measurements, it would be easy to draw a smooth circle/ellipse for each particle if that is needed (I didn't include this in the macro code below that illustrate what I did). Hope this helps, Volko run("Duplicate...", " "); run("8-bit"); setAutoThreshold("Default"); setOption("BlackBackground", true); run("Convert to Mask"); run("Watershed"); run("Set Measurements...", "centroid center fit shape redirect=None decimal=3"); run("Analyze Particles...", "display exclude clear add"); On 29/04/2016 22:21, smithd10 wrote: > Hello, > > I am attempting to size microspheres that I have imaged on TEM. The > microspheres are touching multiple other microspheres and I am having > difficulty segmenting/thresholding the objects to maintain the original size > of the microsphere. I either have the problem of the negative space becoming > the main focus of the particle analysis, or I have thresholded the image so > much that the particle size is no longer accurately measured. > > In other programs, there is the ability to automatically draw circles around > an object based on the circularity of part of that that object. Is that > ability available in ImageJ? > > Thank you. > > <http://imagej.1557.x6.nabble.com/file/n5016288/Pic13.gif> > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Segmenting-Circular-Objects-that-are-touching-multiple-other-objects-tp5016288.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by smithd10
You've cross-posted your request to the ImageJ-forum.
The example-image shows an interesting histogram and I don't understand why a TEM-image comes in indexed (8-bit) colour. You write: "In other programs, there is the ability to automatically draw circles around an object based on the circularity of part of that object." It would be quite interesting to see such results from other programs. Could you please provide an example? Best Herbie :::::::::::::::::::::::::::::::::::::: Am 29.04.16 um 23:21 schrieb smithd10: > Hello, > > I am attempting to size microspheres that I have imaged on TEM. The > microspheres are touching multiple other microspheres and I am having > difficulty segmenting/thresholding the objects to maintain the original size > of the microsphere. I either have the problem of the negative space becoming > the main focus of the particle analysis, or I have thresholded the image so > much that the particle size is no longer accurately measured. > > In other programs, there is the ability to automatically draw circles around > an object based on the circularity of part of that that object. Is that > ability available in ImageJ? > > Thank you. > > <http://imagej.1557.x6.nabble.com/file/n5016288/Pic13.gif> > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Segmenting-Circular-Objects-that-are-touching-multiple-other-objects-tp5016288.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Segmenting Circular Objects that are touching multiple other objects
|
In reply to this post by smithd10
I would approach this with a machine learning approach, like the Trainable Segmentation plugin in Fiji.
Albert > On Apr 29, 2016, at 5:21 PM, smithd10 <[hidden email]> wrote: > > Hello, > > I am attempting to size microspheres that I have imaged on TEM. The > microspheres are touching multiple other microspheres and I am having > difficulty segmenting/thresholding the objects to maintain the original size > of the microsphere. I either have the problem of the negative space becoming > the main focus of the particle analysis, or I have thresholded the image so > much that the particle size is no longer accurately measured. > > In other programs, there is the ability to automatically draw circles around > an object based on the circularity of part of that that object. Is that > ability available in ImageJ? > > Thank you. > > <http://imagej.1557.x6.nabble.com/file/n5016288/Pic13.gif> > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Segmenting-Circular-Objects-that-are-touching-multiple-other-objects-tp5016288.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: Segmenting Circular Objects that are touching multiple other objects
|
In reply to this post by smithd10
I adapted Volko Straub's script to use Jython scripting because it gave me more control and extended an example from the Jython scripting page (link in code). The main point is that one can access the results table. One could make this more complicated by only drawing the well separated particles that are not touching boundaries on the original image. I do this frequently in my particle sizing work. There are also ways to tune the watershed a bit better. I should warn you that Python/Jython is sensitive to indentation. As you can see from the initial part, one needs to import the functions one needs. This helps avoid namespace clashes. Anyway, I just did a small filter based upon circularity and wrote out the equivalent circular diameter values (in pixels) to a .csv.
I typically loop over a series of images, and dump out a feature vector for each blob of interest into a .csv file. I did some very simple processing here. I usually start with a very lenient classifier, dump the data to a .csv and take a look at the data using R. This permits better classification and automated report generation. i have done this with both R/RStudio and lately some with iPython notebooks. Since all these Open Source tools are generally text based, they all (mostly) play well with version control and permit frequently-needed functions to be accumulated in packages to make our work more reproducible. Best wishes as you go down this path. John Minter Eastman Kodak Analytical Sciences Microscopy Lab # procTouchingLatex.py import os import math from ij import IJ, ImagePlus from ij.process import ImageProcessor from ij.process import AutoThresholder from ij.process.AutoThresholder import getThreshold from ij.process.AutoThresholder.Method import Otsu from ij.measure import ResultsTable from ij.plugin.filter import EDM # I use environment variables and store images and reports is predictable directories. # To test, you can just make a path to a working directory. Note that Windows, Mac # and Linux will let you use a forward slash... homDir = os.environ['HOME'] relPrj = "/dat/images/IJ/" wrkDir = homDir + relPrj # put main config parameters at the top.... minCirc = 0.840 # see num 5 below iDigits = 4 # for rounding sampID = 'latex-TEM' bVerbose = False sImgPath = wrkDir + sampID + '.gif' sCsvPath = wrkDir + sampID + ".csv" ori = ImagePlus(sImgPath) ori.show() # Make a copy of the original image, doing the conversion to 8 bit: # adapted from IJ Jython Scripting page http://imagej.net/Jython_Scripting wrk = ori.createImagePlus() ip = ori.getProcessor().duplicate().convertToByteProcessor() wrk.setProcessor("Latex", ip) stats = wrk.getStatistics() his = stats.histogram # 2 - Apply a threshold: only zeros and ones # # This is from Otsu, set manually thr = AutoThresholder().getThreshold(Otsu, his) if bVerbose: print(thr) ip.setThreshold(0, thr, ImageProcessor.NO_LUT_UPDATE) # Call the Thresholder to convert the image to a mask IJ.run(wrk, "Convert to Mask", "") # 3 - Apply watershed # Create and run new EDM object, which is an Euclidean Distance Map (EDM) # and run the watershed on the ImageProcessor: EDM().toWatershed(ip) # 4 - Show the watershed image wrk.show() # 5 set up the measurements and compute them. Writes them to the results # table. Compare particles 1, 75,76, and 77 (partially extracted spheres) to the others. # "Good" spheres have a circularity of > about 0.83 IJ.run(wrk, "Set Measurements...", "area centroid center fit shape redirect=None decimal=3") IJ.run(wrk, "Analyze Particles...", "display exclude clear add") # 6 - Get the results table and compute the ECD for the "Good" Particles rt = ResultsTable().getResultsTable() lArea = rt.getColumn(rt.getColumnIndex("Area")) lCirc = rt.getColumn(rt.getColumnIndex("Circ.")) nPart = len(lArea) partID = [] partECD = [] k=0 for i in range(nPart): area = lArea[i] circ = lCirc[i] ecd = 2.0 * math.sqrt(area/math.pi) if circ > minCirc: partID.append(i+1) partECD.append(round(ecd, iDigits)) if bVerbose: print(i+1, ecd) # write the output file as .csv f=open(sCsvPath, 'w') strLine = 'part, ecd.px\n' f.write(strLine) for k in range(len(partECD)): strLine = "%d, %.5f\n" % (partID[k], partECD[k] ) f.write(strLine) f.close() |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

