counting cells in multiclour fluorescence images
counting cells in multiclour fluorescence images
|
Dear all,
thanks to all of you for your suggestions towards a solution for what I think is a really 'real life' application. Of course the sample image is only a small sample of the hundreds of images I would like to process objectively and without the stress of deciding whether cells belong to one class or the other. I'd imagine that many of you are facing similar problems and most of you are more fluent in constructing a working routine. The main problems start with a real good solution for separating touching nuclei. Watersheding as provided in ImageJ does not work too reliably in many instances. Does anybody know of an alternative routine?? If one could make a wish, a solution which implements user knowledge would be helpful. Something along the lines: this particle looks to big, to indented, not round enough to be a single nucleus, so lets split it. If nuclei were separated correctly, the problem of attributing a piece of cytoplasm to this nucleus that does not belong to another cell is quite a big one. Theoretically (or better wishful thinking), I would imagine a solution which starts to look in the surrounding of the nucleus for the same staining pattern and extends that region until it encounters a rivaling cell, which also grows its cytoplasm or until it reaches the plasma membrane where the staining sometimes fades away or drops apruptely. So far I have been trying to use an approach influenced by suggestions and copious help from G. Landini and J. Mutterer to generate 'annuli' around the identified nuclei. However, if the cells are closely spaced, the area around a particular nucleus is splitted, in the end producing more particles than cells. Another problem obviously comes from processes of neighbouring cells running close to a nucleus of a cell belonging to another class (i.e. not stained). This could of course screw up the classification of cells quite substantially. Taken together, I still have not worked out (as an image analysis dummy, may never will) a satisfying solution (so generations of graduate students will suffer from trying to count cells manually) and would really appreciate help from the ImageJ community- Below are snapshots (along with the original starting image in tif.format) of my current (unsatisfying) analysis approach). Of course I am aware that all of this is asking very much. However, faced with 'real life' problems, which cry for an image analyis solution, I would appreciate any suggestions, which lead to a working solution. Thanks for any input! Sincerely Matthias 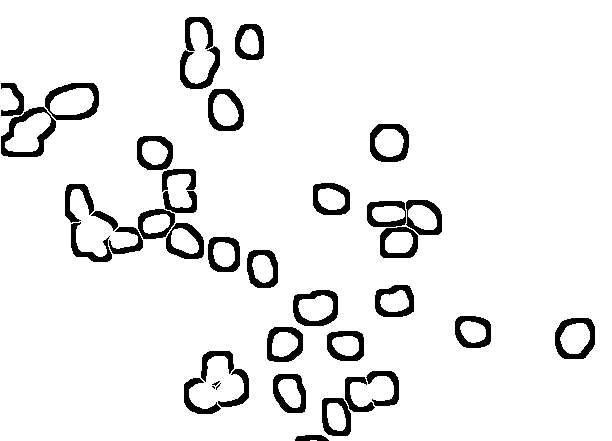 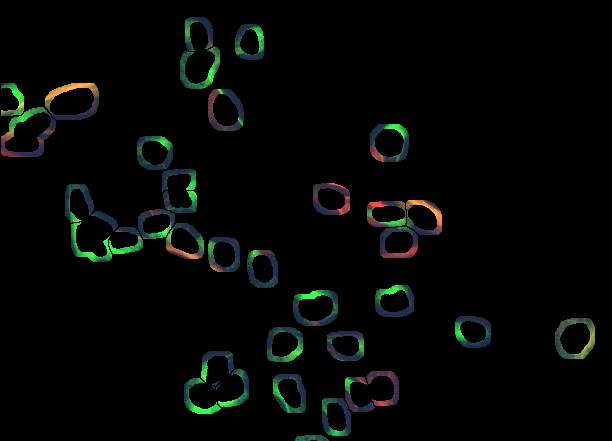 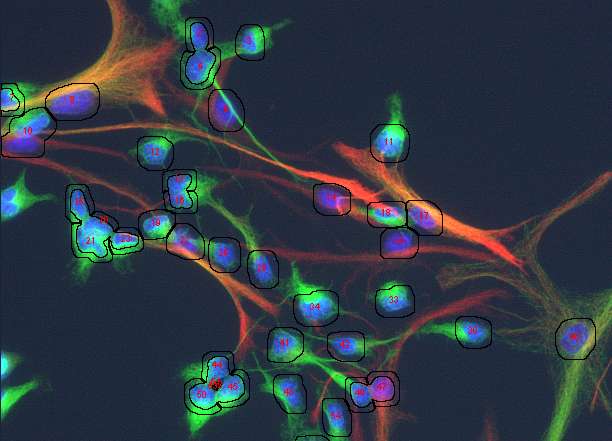
|
Re: counting cells in multiclour fluorescence images
|
On Wednesday 19 November 2008, Matthias Kirsch wrote:
> The main problems start with a real good solution for separating touching > nuclei. Watersheding as provided in ImageJ does not work too reliably in > many instances. It does do what is intended (ie watershed separation). The problem is that the nuclei clusters do not always give clues as to where the separating lines should be placed. > Does anybody know of an alternative routine?? If one could > make a wish, a solution which implements user knowledge would be helpful. There is a Watershed plugin from Daniel Sage that can process greyscale images and might be useful (search in goolge). Be aware, however, that if you use that plugin and set up a threshold *within the plugin*, there is a problem obtaining 8-connected basins (i.e. 4-connected watershed lines). I have been in contact with Daniel in the past about this problem, but I do not think it has been solved yet. If you use the whole greyscale space (i.e. minLevel=0, maxLevel=255) the plugin works fine, but you might be segmenting also areas you do not need, so some workaround is necessary (i.e. set all background areas to 0 or someting like that). So, to summarise, if your set min and max levels, the plugin will produce watershed lines, but you cannot isolate separated cells with the particle analyzer or the particles8 plugins. If you set levels to 0 and 255 the plugin works fine. > generate 'annuli' around > the identified nuclei. However, if the cells are closely spaced, the area > around a particular nucleus is splitted, in the end producing more > particles than cells. You can still select those many fragments which are continous to a particular nucleus and count them as one. > Another problem obviously comes from processes of neighbouring cells > running close to a nucleus of a cell belonging to another class (i.e. not > stained). This could of course screw up the classification of cells quite > substantially. I guess that is not a problem that can be solved in 2D images. Cells overlap and one has lost 1 dimension, so which part of the cytplsasm belongs to which level? One solution is to look at cells which are isolated, but in itself is in a way introducing bias in the analysis as cell contacts must surely change the way they behave. When separation is not possible, you can still look at the behavious of clustered cells vs isolated cells. That might give you some information too. I hope it helps Gabriel |
|
In reply to this post by Matthias Kirsch
Hallo, possibly a bit late some add. hints for the problem stated:
1. Segment in blue the nuclei, connected nuclei should be cutted by hand 2. Skeletonize the background of the nuclei The result might look a bit chaotic, but using for particle analysis the regions, which correspond to each nucleus, redirected to the red and green channel (total intensity) will deliver you per nucleus the amount of signal in the regions defined by the skeleton. If that is not good enough (the skeleton is not dependent on cell area) you can try around with watershed in the segmented intensity image of the original. However, as far as you can easily detect the nuclei it is always possible to divide the interesting area (background of nuceli (NOT nucei), cellular area into corresponding areas which might deliver you a better estimate of intensities compared to your "near-vicinity" trials! Generate from the skeleton areas rois and measure total intensity in red and green channel. Regards Karsten Am 19.11.2008 um 20:01 schrieb Matthias Kirsch: > Dear all, > > thanks to all of you for your suggestions towards a solution for > what I > think is a really 'real life' application. Of course the sample > image is > only a small sample of the hundreds of images I would like to process > objectively and without the stress of deciding whether cells belong > to one > class or the other. I'd imagine that many of you are facing similar > problems > and most of you are more fluent in constructing a working routine. > The main problems start with a real good solution for separating > touching > nuclei. Watersheding as provided in ImageJ does not work too > reliably in > many instances. Does anybody know of an alternative routine?? If one > could > make a wish, a solution which implements user knowledge would be > helpful. > Something along the lines: this particle looks to big, to indented, > not > round enough to be a single nucleus, so lets split it. > > If nuclei were separated correctly, the problem of attributing a > piece of > cytoplasm to this nucleus that does not belong to another cell is > quite a > big one. Theoretically (or better wishful thinking), I would imagine a > solution which starts to look in the surrounding of the nucleus for > the same > staining pattern and extends that region until it encounters a > rivaling > cell, which also grows its cytoplasm or until it reaches the plasma > membrane > where the staining sometimes fades away or drops apruptely. So far > I have > been trying to use an approach influenced by suggestions and copious > help > from G. Landini and J. Mutterer to generate 'annuli' around the > identified > nuclei. However, if the cells are closely spaced, the area around a > particular nucleus is splitted, in the end producing more particles > than > cells. > Another problem obviously comes from processes of neighbouring cells > running > close to a nucleus of a cell belonging to another class (i.e. not > stained). > This could of course screw up the classification of cells quite > substantially. > > Taken together, I still have not worked out (as an image analysis > dummy, may > never will) a satisfying solution (so generations of graduate > students will > suffer from trying to count cells manually) and would really > appreciate help > from the ImageJ community- > > Below are snapshots (along with the original starting image in > tif.format) > of my current (unsatisfying) analysis approach). > > > > Of course I am aware that all of this is asking very much. However, > faced > with 'real life' problems, which cry for an image analyis solution, > I would > appreciate any suggestions, which lead to a working solution. > > > Thanks for any input! > > Sincerely > > Matthias http://n2.nabble.com/file/n1520189/2_subset%2Bof%2Bannuli.jpg > http://n2.nabble.com/file/n1520189/3.jpg > http://n2.nabble.com/file/n1520189/4_final.jpg > -- > View this message in context: http://n2.nabble.com/counting-cells-in-multiclour-fluorescence-images-tp1520189p1520189.html > Sent from the ImageJ mailing list archive at Nabble.com. |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

