counting cells in multiclour fluorescence images
counting cells in multiclour fluorescence images
|
Dear all,
I am trying to find a way to count cells expressing one or the other or both proteins in cultured cells (see e.g. the attached example). There are green cells, red cells and yellow cells and I would like to have a (semi)-automatic routine with which this can be done objectively. Basically the cells can be identified by their nuclei (in the blue channel), but I do not know how to count only the nuclei which have a red, green or yellow cytoplasm associated with them. As there probably is no way to reliably find the outline of a cell with any accuracy (however, maybe maybe one has an ingenious approach), it may be sufficient to look in the immediate vicinity of the nuclei (e.g. in a ring some pixels wide around the nuclei -user selectable preferrentially), if this area is red, green or yellow. I would appreciate if someone could help to solve this problem. Thanx Matthias 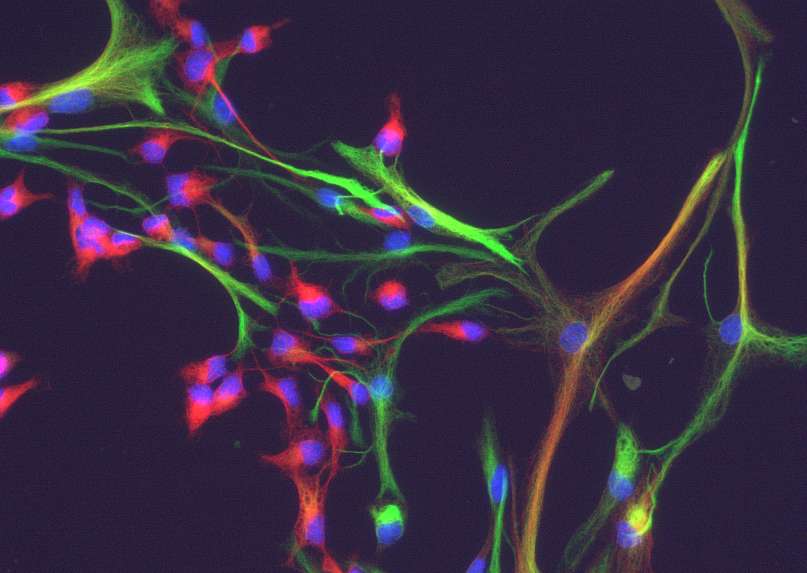 |
Re: counting cells in multiclour fluorescence images
|
This is a particularly interesting problem, since in a single 3-color
image, the yellow arises from the combination of red and green. Thus, it is possible to separate the color channels to get a red signal, a green one, and the blue one. However, both the red and the green channels also carry the information for the yellow cells. Here's a thought: You can subtract green from red to get those cells that are only red, and not yellow. Similarly, you can subtract red from green to get those that are only green. Then, you can add red and green to get an enhancement of the yellow signal. If your signals are consistent, you can do this in a macro, and then assign threshold values, and use the analyze particles routine. Best of luck. Joel Date sent: Sun, 16 Nov 2008 12:58:33 -0800 Send reply to: ImageJ Interest Group <[hidden email]> From: Matthias Kirsch <[hidden email]> Subject: counting cells in multiclour fluorescence images To: [hidden email] > Dear all, > > I am trying to find a way to count cells expressing one or the other or both > proteins in cultured cells (see e.g. the attached example). There are green > cells, red cells and yellow cells and I would like to have a > (semi)-automatic routine with which this can be done objectively. Basically > the cells can be identified by their nuclei (in the blue channel), but I do > not know how to count only the nuclei which have a red, green or yellow > cytoplasm associated with them. As there probably is no way to reliably find > the outline of a cell with any accuracy (however, maybe maybe one has an > ingenious approach), it may be sufficient to look in the immediate vicinity > of the nuclei (e.g. in a ring some pixels wide around the nuclei -user > selectable preferrentially), if this area is red, green or yellow. > I would appreciate if someone could help to solve this problem. > > Thanx > Matthias http://n2.nabble.com/file/n1507033/cells.jpg > > > > -- > View this message in context: http://n2.nabble.com/counting-cells-in-multiclour-fluorescence-images-tp1507033p1507033.html > Sent from the ImageJ mailing list archive at Nabble.com. Joel B. Sheffield, Ph.D Department of Biology Temple University Philadelphia, PA 19122 Voice: 215 204 8839 e-mail: [hidden email] URL: http://astro.temple.edu/~jbs |
Re: counting cells in multiclour fluorescence images
|
In reply to this post by Matthias Kirsch
I've been playing with some semi-automated cell identification and put the macro so far below. It's based on automated cell segmentation in high content screening where a nucleic acid stain (DARQ5 in our case) is used to first identify the nucleus (DNA really bright) then the cytoplasm (RNA being fainter). You can kind of generate a similar type of image by splitting your RGB, dividing the red and green by 2 to make them fainter and adding the red green and blue back together.
The macro uses two thresholds one for nucleus and one for cytoplasm which are here in the text (a better tweak would be to pull the upper and lower bounds from the user defined threshold). It uses particle detection to find nuclei then maxima detection within the lower threshold bound to find the cytoplasm. Once it's identified the cells and put them in the ROI manager then you can go crazy with the analysis. http://www.macbiophotonics.ca/images/cellSegmentation.jpg Hope this helps. Regards, Tony cytoThresh = 4; nucThresh=35; origID=getImageID(); run("Duplicate...", "title=nuc"); run("Gaussian Blur...", "sigma=1"); setThreshold(nucThresh, 255); run("Convert to Mask"); run("Watershed"); run("Ultimate Points"); run("Multiply...", "value=255"); run("Dilate"); run("Dilate"); nucID=getImageID(); selectImage(origID); run("Duplicate...", "title=cyto"); cytoID=getImageID(); run("Gaussian Blur...", "sigma=1"); run("Divide...", "value=2"); imageCalculator("Add", cytoID,nucID); setThreshold(cytoThresh , 255); run("Find Maxima...", "noise=50 output=[Segmented Particles] above"); segmentedID=getImageID(); setThreshold(1, 25500); run("Analyze Particles...", "size=150-25000 circularity=0.00-1.00 show=Outlines clear summarize add"); maskID=getImageID(); selectImage(cytoID); close(); selectImage(nucID); close(); selectImage(maskID); close(); selectImage(segmentedID); close(); selectImage(origID); setOption("Show All",true); roiManager("Measure"); Tony J. Collins, Ph.D. McMaster Biophotonics Facility Dept. Biochemistry and Biomedical Sciences HSC 4H21A McMaster University, Hamilton, ON, L8N 3Z5 [hidden email] www.macbiophotonics.ca > -----Original Message----- > From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of > Matthias Kirsch > Sent: Sunday, November 16, 2008 3:59 PM > To: [hidden email] > Subject: counting cells in multiclour fluorescence images > > Dear all, > > I am trying to find a way to count cells expressing one or the other or > both > proteins in cultured cells (see e.g. the attached example). There are > green > cells, red cells and yellow cells and I would like to have a > (semi)-automatic routine with which this can be done objectively. > Basically > the cells can be identified by their nuclei (in the blue channel), but > I do > not know how to count only the nuclei which have a red, green or yellow > cytoplasm associated with them. As there probably is no way to reliably > find > the outline of a cell with any accuracy (however, maybe maybe one has > an > ingenious approach), it may be sufficient to look in the immediate > vicinity > of the nuclei (e.g. in a ring some pixels wide around the nuclei -user > selectable preferrentially), if this area is red, green or yellow. > I would appreciate if someone could help to solve this problem. > > Thanx > Matthias http://n2.nabble.com/file/n1507033/cells.jpg > > > > -- > View this message in context: http://n2.nabble.com/counting-cells-in- > multiclour-fluorescence-images-tp1507033p1507033.html > Sent from the ImageJ mailing list archive at Nabble.com. |
Re: counting cells in multiclour fluorescence images
|
On Tuesday 18 November 2008 20:47:27 Tony Collins wrote:
> I've been playing with some semi-automated cell identification and put the > macro so far below. It's based on automated cell segmentation in high > content screening where a nucleic acid stain (DARQ5 in our case) is used to > first identify the nucleus (DNA really bright) then the cytoplasm (RNA > being fainter). You can kind of generate a similar type of image by > splitting your RGB, dividing the red and green by 2 to make them fainter > and adding the red green and blue back together. > > The macro uses two thresholds one for nucleus and one for cytoplasm which > are here in the text (a better tweak would be to pull the upper and lower > bounds from the user defined threshold). > > It uses particle detection to find nuclei then maxima detection within the > lower threshold bound to find the cytoplasm. Once it's identified the cells > and put them in the ROI manager then you can go crazy with the analysis. > > http://www.macbiophotonics.ca/images/cellSegmentation.jpg The problem with this type of approach is that the cytoplasms are all different sizes, so one is not comparing the same thing. E.g. is it a fair comparison between cell 30 and cell 12?. What about thresholding the blue channel (nuclei), make sure that each nucleus is separated (one could run the Binary Watershed for this) and then do a "Dilate-no-merge" of the nuclei (let's say 3 or 4 iterations) (a plugin for this is available in the Morphology collection). Then subtract the nucleus mask from this "dilated-not-merged" mask (which should leave binary "rings" that have an upper bound more comparable between cells). Finally do a logical AND of the ring mask and the original and analyse the contribution of the red and green channels. Cheers Gabriel |
Re: counting cells in multiclour fluorescence images
|
In reply to this post by Tony Collins-4
Dear Tony,
thanks for your suggesstions. I have tried your macro and it produces weird results in my hands and I do not know where the problem is? Both the input image and the one processed with your macro (used directly as you have put it in your e-mail to me) are attached. Cheers Matthias On Tue, Nov 18, 2008 at 9:49 PM, Tony Collins-4 (via Nabble) <[hidden email]> wrote: I've been playing with some semi-automated cell identification and put the macro so far below. It's based on automated cell segmentation in high content screening where a nucleic acid stain (DARQ5 in our case) is used to first identify the nucleus (DNA really bright) then the cytoplasm (RNA being fainter). You can kind of generate a similar type of image by splitting your RGB, dividing the red and green by 2 to make them fainter and adding the red green and blue back together. |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

