deconvolution problem
|
Hi,
Has anyone tried the deconvolution lab with ImageJ, with the dataset of fluorescent beads, which is provided by http://bigwww.epfl.ch/deconvolution/?p=bars ? I have tried, but I cannot get the result as they shows, I don't know why. Is there anyone can help? Thanks, Amy -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Amy
My suggestion is to try DeconvolutionLab with the following settings: Set the Algorithm to Richardson Lucy, set the iterations to 100, make sure you set the PSF to the correct data set, then check "Flip PSF quadrants" (if the PSF is centered). I find you have to be careful and make sure the image you want to deconvolve is focused before opening the plugin. When the deconvolution starts watch the output in the log window carefully and verify it is loading the correct input and psf and flipping the psf if needed. With these settings you should be able to get an OK result, and at least confirm it is working. Then you could try Richardson-Lucy with TV regularization and/or Thresholded Landweber. Theoretically these algorithms should give better results, at least with noisy images but might require more adjusting of advanced paramaters. Brian On Mon, Jan 14, 2013 at 3:41 PM, Amy Cao <[hidden email]> wrote: > Hi, > > Has anyone tried the deconvolution lab with ImageJ, with the dataset of > fluorescent beads, which is provided by > http://bigwww.epfl.ch/deconvolution/?p=bars ? I have tried, but I cannot > get the result as they shows, I don't know why. Is there anyone can help? > > Thanks, > > Amy > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Brian,
Thank you so much for your suggestion, I have made it according to your suggestion. I think my problem is I did not flip PSF. But I don't understand why I should flip PSF, do you have some idea on it? Thanks, Amy On Mon, Jan 14, 2013 at 8:27 PM, Brian Northan <[hidden email]> wrote: > Hi Amy > > My suggestion is to try DeconvolutionLab with the following settings: > > Set the Algorithm to Richardson Lucy, set the iterations to 100, make > sure you set the PSF to the correct data set, then check "Flip PSF > quadrants" (if the PSF is centered). I find you have to be careful > and make sure the image you want to deconvolve is focused before > opening the plugin. When the deconvolution starts watch the output in > the log window carefully and verify it is loading the correct input > and psf and flipping the psf if needed. > > With these settings you should be able to get an OK result, and at > least confirm it is working. Then you could try Richardson-Lucy with > TV regularization and/or Thresholded Landweber. Theoretically these > algorithms should give better results, at least with noisy images but > might require more adjusting of advanced paramaters. > > Brian > > On Mon, Jan 14, 2013 at 3:41 PM, Amy Cao <[hidden email]> wrote: > > Hi, > > > > Has anyone tried the deconvolution lab with ImageJ, with the dataset of > > fluorescent beads, which is provided by > > http://bigwww.epfl.ch/deconvolution/?p=bars ? I have tried, but I cannot > > get the result as they shows, I don't know why. Is there anyone can help? > > > > Thanks, > > > > Amy > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Amy
Yes. The equations in the deconvolution consider the center of the PSF to be 0,0,0. While on the other hand in an image the center is ussually at width/2, height/2, depth/2. So flip PSF moves the coordinate system to where the algorithm needs it to be. Brian -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Brian,
Is that mean all the algorithms in Deconvolution Lab require the center of the PSF to be 0,0,0? Sorry about this simple question, because I am not doing the algorithms things, and I always confused about that. Is it possible for you maybe to recommend some papers about this deconvolution so that I can get some ideas about it? Thank you so much, Jianwei Cao On Thu, Jan 17, 2013 at 6:50 AM, Brian Northan <[hidden email]> wrote: > Hi Amy > > Yes. The equations in the deconvolution consider the center of the > PSF to be 0,0,0. While on the other hand in an image the center is > ussually at width/2, height/2, depth/2. So flip PSF moves the > coordinate system to where the algorithm needs it to be. > > Brian > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Amy
Yes as far as I know all the algorithms in Deconvolution Lab need the PSF to be shifted. In terms of papers if you want something general I would recomend the following. Biggs, S. C., “A practical guide to deconvolution of fluorescence microscope imagery,” Microscopy Today 18(1), 10-14 (2010). In terms of specific details for DeconvolutionLab... they implement a few algorithms. Richardson Lucy is the basic one. Richardson Lucy with TV Regularization and Thresholded Landweber are more advanced. They should produce better results but have more complicated input parameters. I tried them a couple of times before but didn't get the result I expected. Though I personally haven't had a chance to work with those algorithms much and figure out the right settings. Maybe someone else has more experience with them?? You should keep asking around. The references for those algorithms are below.... Dey, N., Bland-Feraud L., Zimmer, C., Roux, P., Kam, Z., Olivo-Marin, J.-C., and Zerubia, J., “Richardson-Lucy algorithm with total variation regularization for 3D confocal microscope deconvolution,” Microscopy Research and Technique, 69, 260-266 (2006). Vonesch, C. and Unser, M., “A fast thresholded Landweber algorithm for wavelet-regularized multidimensional deconvolution,” IEEE transactions on image processing, 17(4), 539-549 (2008). On Fri, Jan 18, 2013 at 3:43 PM, Amy Cao <[hidden email]> wrote: > Hi Brian, > > Is that mean all the algorithms in Deconvolution Lab require the center of > the PSF to be 0,0,0? > > Sorry about this simple question, because I am not doing the algorithms > things, and I always confused about that. Is it possible for you maybe to > recommend some papers about this deconvolution so that I can get some ideas > about it? > > Thank you so much, > > Jianwei Cao > > On Thu, Jan 17, 2013 at 6:50 AM, Brian Northan <[hidden email]> wrote: > >> Hi Amy >> >> Yes. The equations in the deconvolution consider the center of the >> PSF to be 0,0,0. While on the other hand in an image the center is >> ussually at width/2, height/2, depth/2. So flip PSF moves the >> coordinate system to where the algorithm needs it to be. >> >> Brian >> >> -- >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html >> > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Brian,
Thanks for the recommendation, it really helps! I would keep on learning. Best wishes, Amy Cao On Fri, Jan 18, 2013 at 4:01 PM, Brian Northan <[hidden email]> wrote: > Hi Amy > > Yes as far as I know all the algorithms in Deconvolution Lab need the > PSF to be shifted. > > In terms of papers if you want something general I would recomend the > following. > > Biggs, S. C., “A practical guide to deconvolution of fluorescence > microscope imagery,” Microscopy Today 18(1), 10-14 (2010). > > In terms of specific details for DeconvolutionLab... they implement a > few algorithms. Richardson Lucy is the basic one. Richardson Lucy > with TV Regularization and Thresholded Landweber are more advanced. > They should produce better results but have more complicated input > parameters. I tried them a couple of times before but didn't get the > result I expected. Though I personally haven't had a chance to work > with those algorithms much and figure out the right settings. Maybe > someone else has more experience with them?? You should keep asking > around. The references for those algorithms are below.... > > Dey, N., Bland-Feraud L., Zimmer, C., Roux, P., Kam, Z., Olivo-Marin, > J.-C., and Zerubia, J., “Richardson-Lucy algorithm with total > variation regularization for 3D confocal microscope deconvolution,” > Microscopy Research and Technique, 69, 260-266 (2006). > > Vonesch, C. and Unser, M., “A fast thresholded Landweber algorithm for > wavelet-regularized multidimensional deconvolution,” IEEE transactions > on image processing, 17(4), 539-549 (2008). > > On Fri, Jan 18, 2013 at 3:43 PM, Amy Cao <[hidden email]> wrote: > > Hi Brian, > > > > Is that mean all the algorithms in Deconvolution Lab require the center > of > > the PSF to be 0,0,0? > > > > Sorry about this simple question, because I am not doing the algorithms > > things, and I always confused about that. Is it possible for you maybe to > > recommend some papers about this deconvolution so that I can get some > ideas > > about it? > > > > Thank you so much, > > > > Jianwei Cao > > > > On Thu, Jan 17, 2013 at 6:50 AM, Brian Northan <[hidden email]> > wrote: > > > >> Hi Amy > >> > >> Yes. The equations in the deconvolution consider the center of the > >> PSF to be 0,0,0. While on the other hand in an image the center is > >> ussually at width/2, height/2, depth/2. So flip PSF moves the > >> coordinate system to where the algorithm needs it to be. > >> > >> Brian > >> > >> -- > >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > >> > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by bnorthan
Hi Brian
I had the same question with Amy, too. Thank you for the suggestion about flipping the PSF. But I have other problems using the "Batch" module. I tried to load the image stacks like showed in the photo: <http://imagej.1557.x6.nabble.com/file/n5015763/Forum.png> But failed to run deconvolution even all the images were 2D tiff. And the dimension of the PSF was smaller than the image stacks. Is there any file format limitation for the "Batch" module? Thanks, Mirrownight bnorthan wrote > Hi Amy > > Yes as far as I know all the algorithms in Deconvolution Lab need the > PSF to be shifted. > > In terms of papers if you want something general I would recomend the > following. > > Biggs, S. C., “A practical guide to deconvolution of fluorescence > microscope imagery,” Microscopy Today 18(1), 10-14 (2010). > > In terms of specific details for DeconvolutionLab... they implement a > few algorithms. Richardson Lucy is the basic one. Richardson Lucy > with TV Regularization and Thresholded Landweber are more advanced. > They should produce better results but have more complicated input > parameters. I tried them a couple of times before but didn't get the > result I expected. Though I personally haven't had a chance to work > with those algorithms much and figure out the right settings. Maybe > someone else has more experience with them?? You should keep asking > around. The references for those algorithms are below.... > > Dey, N., Bland-Feraud L., Zimmer, C., Roux, P., Kam, Z., Olivo-Marin, > J.-C., and Zerubia, J., “Richardson-Lucy algorithm with total > variation regularization for 3D confocal microscope deconvolution,” > Microscopy Research and Technique, 69, 260-266 (2006). > > Vonesch, C. and Unser, M., “A fast thresholded Landweber algorithm for > wavelet-regularized multidimensional deconvolution,” IEEE transactions > on image processing, 17(4), 539-549 (2008). > > On Fri, Jan 18, 2013 at 3:43 PM, Amy Cao < > amycao723@ > > wrote: >> Hi Brian, >> >> Is that mean all the algorithms in Deconvolution Lab require the center >> of >> the PSF to be 0,0,0? >> >> Sorry about this simple question, because I am not doing the algorithms >> things, and I always confused about that. Is it possible for you maybe to >> recommend some papers about this deconvolution so that I can get some >> ideas >> about it? >> >> Thank you so much, >> >> Jianwei Cao >> >> On Thu, Jan 17, 2013 at 6:50 AM, Brian Northan < > bnorthan@ > > wrote: >> >>> Hi Amy >>> >>> Yes. The equations in the deconvolution consider the center of the >>> PSF to be 0,0,0. While on the other hand in an image the center is >>> ussually at width/2, height/2, depth/2. So flip PSF moves the >>> coordinate system to where the algorithm needs it to be. >>> >>> Brian >>> >>> -- >>> ImageJ mailing list: http://imagej.nih.gov/ij/list.html >>> >> >> -- >> ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- View this message in context: http://imagej.1557.x6.nabble.com/deconvolution-problem-tp5001407p5015763.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by bnorthan
Hi Brian
I had the same question with Amy, too. Thank you for the suggestion about flipping the PSF. But I have other problems using the "Batch" module. I tried to load the image stacks like showed in the photo: 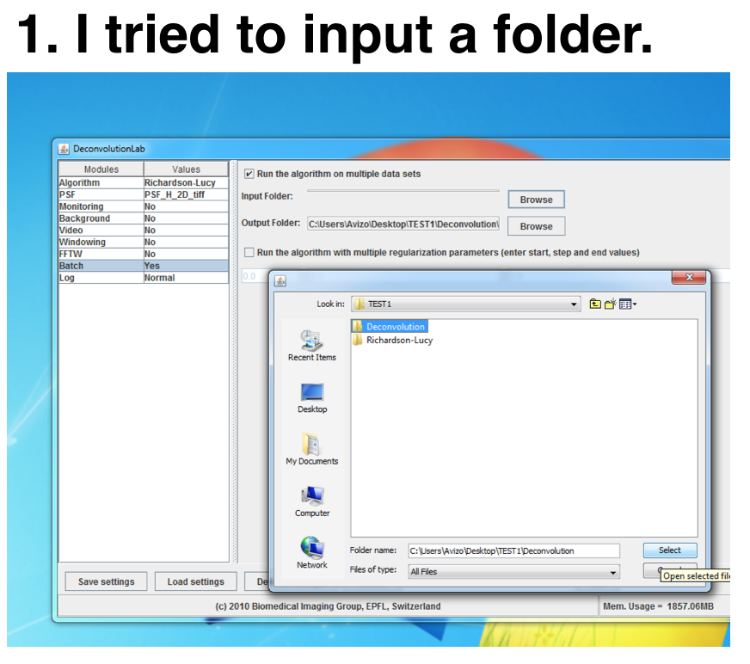 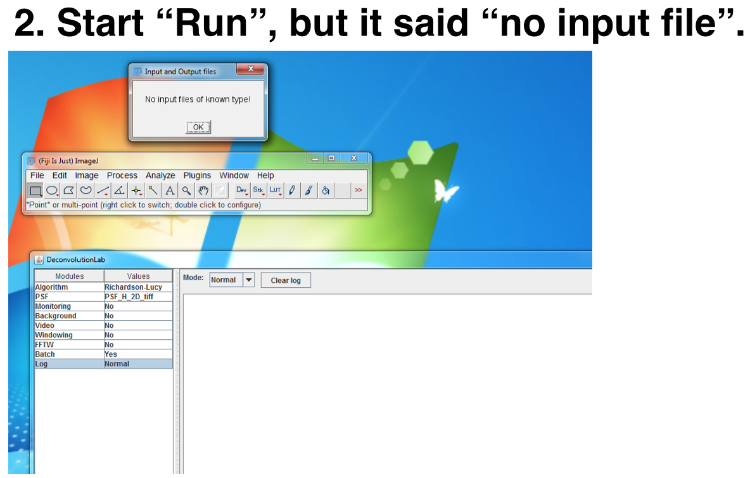 I failed to run deconvolution even all the images were 2D tiff. And the dimension of the PSF was smaller than the image stacks. Is there any file format limitation for the "Batch" module? Thanks, Mirrownight |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

