object-oriented segmentation
object-oriented segmentation
|
Hi,
Does anyone know a plugin or a method to perform object-oriented segmentation in ImageJ? I have 2 images of the same sample. Image 1 contains a nuclear staining, while image 2 contains a cytoplasmic staining of the same cells. I would like to segment image 2 in such a way that every ROI contains one nucleus from image 1. What I was trying to do: Segment image 1, make ROIs, transfer ROIs to image 2, and expand ROIs until they either touch another ROI or until they touch the edge of cytoplasmic staining. But the expanding part does not stop when touching another ROI or the edge of cytoplasmic staining. Another way could be some sort of ROI seeded watershed, but I have no idea to accomplish this. Can anyone please help me out? Thanks, Michiel |
|
Dear Michiel,
You could try: Take your segmented image and process > Binary > Voronoi. Next take the resulting image and Threshold this image with settings that show all the lines. Run Analyze > Analyze Particles with only Add to Manager selected. Or Take your original image, Process > Find Maxima and select as Output type: Segmented Particles. Again take the results image and run Analyze > Analyze Particles with only Add to Manager selected. Hope this works. Best wishes. Kees Dr Ir K.R. Straatman Senior Experimental Officer Centre for Core Biotechnology Services University of Leicester http://www.le.ac.uk/biochem/microscopy/home.html -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: 11 December 2012 08:46 To: [hidden email] Subject: object-oriented segmentation Hi, Does anyone know a plugin or a method to perform object-oriented segmentation in ImageJ? I have 2 images of the same sample. Image 1 contains a nuclear staining, while image 2 contains a cytoplasmic staining of the same cells. I would like to segment image 2 in such a way that every ROI contains one nucleus from image 1. What I was trying to do: Segment image 1, make ROIs, transfer ROIs to image 2, and expand ROIs until they either touch another ROI or until they touch the edge of cytoplasmic staining. But the expanding part does not stop when touching another ROI or the edge of cytoplasmic staining. Another way could be some sort of ROI seeded watershed, but I have no idea to accomplish this. Can anyone please help me out? Thanks, Michiel -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
Dear Kees,
Thanks for the quick response. I am afraid this is not exactly what I am looking for. The way I interpreted your two methods, both “Vonoroi” as well as “Find Maxima” on the nuclear signal do not take the cytoplasmic signal into account. This way, the ROIs that are created are much bigger than the cytoplasmic signal. Or am I misinterpreting your advice? I will be a bit more explanatory in my question. This time I included two images, one with nuclear signal (Hoechst) and one with cytoplasmic signal (Actinin), processed with a different program. The segmentation is not perfect, but this is more or less what I would like to do with ImageJ. Unfortunately not all of the cells have Actinin, on average about 10% of the nuclei do not have Actinin. I am only interested in the cells that do contain Actinin. Two of the parameters I am looking for are cellular area (either pixels or square um) and average intensity per cell. Eventually I would also like to transfer the ROIs to a third channel image for quantification of a third color. And ultimately I would like to automate this to batch process hundreds of images like this. Is there a way to use “Vonoroi” or “Find Maxima” in such a way that the edges of the ROIs end with the edges of the cytoplasmic signal (Actinin)? Or is there a different way to obtain these ROIs? Thanks again, Michiel 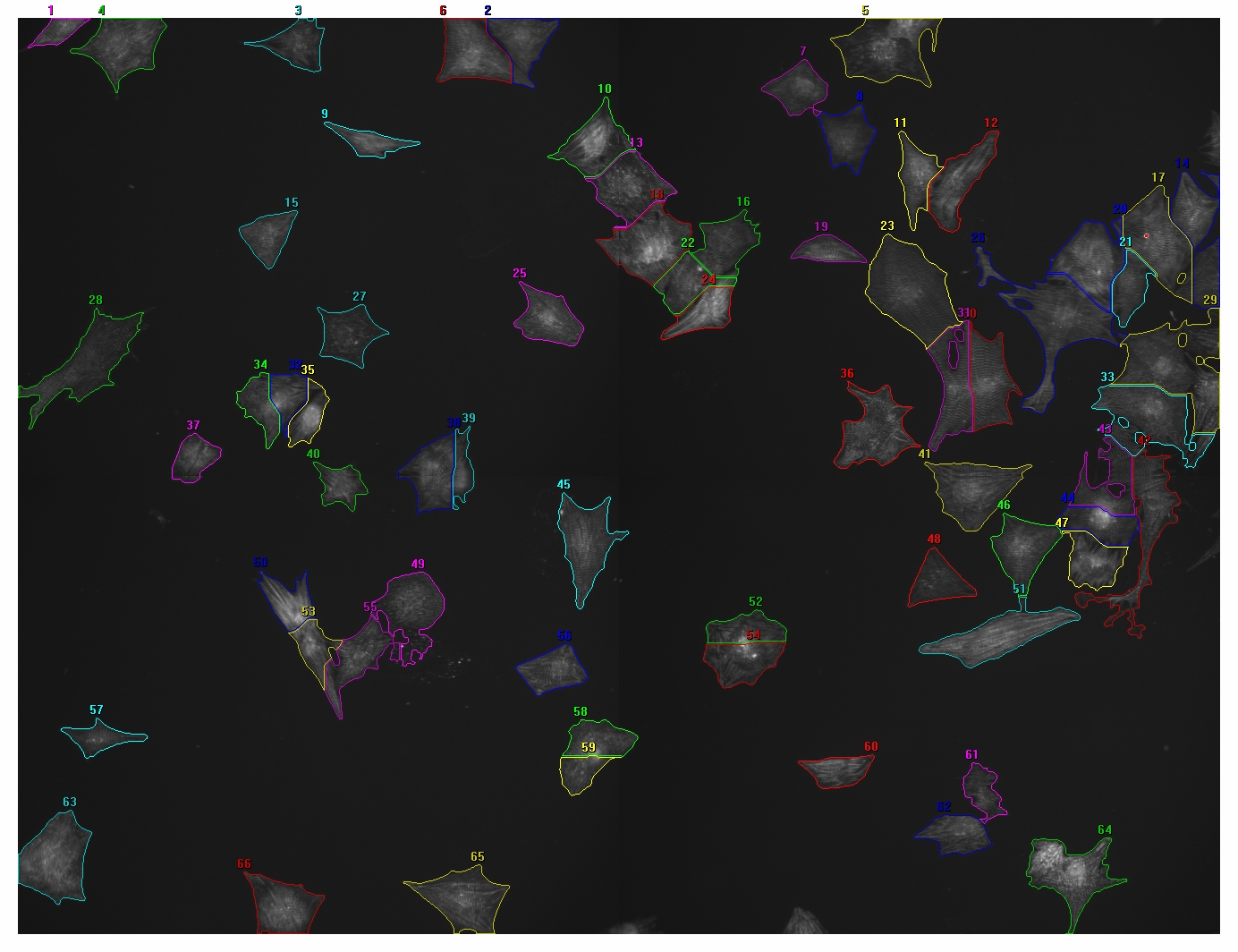 
|
Re: object-oriented segmentation
|
Michiel,
Have you had a look at the "objects based" colocalization available in JACoP? Manuscript found here: http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1365-2818.2006.01706.x Plugin description found here: http://imagej.nih.gov/ij/plugins/track/jacop.html Dave -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: Tuesday, December 11, 2012 6:30 AM To: [hidden email] Subject: Re: object-oriented segmentation Dear Kees, Thanks for the quick response. I am afraid this is not exactly what I am looking for. The way I interpreted your two methods, both “Vonoroi” as well as “Find Maxima” on the nuclear signal do not take the cytoplasmic signal into account. This way, the ROIs that are created are much bigger than the cytoplasmic signal. Or am I misinterpreting your advice? I will be a bit more explanatory in my question. This time I included two images, one with nuclear signal (Hoechst) and one with cytoplasmic signal (Actinin), processed with a different program. The segmentation is not perfect, but this is more or less what I would like to do with ImageJ. Unfortunately not all of the cells have Actinin, on average about 10% of the nuclei do not have Actinin. I am only interested in the cells that do contain Actinin. Two of the parameters I am looking for are cellular area (either pixels or square um) and average intensity per cell. Eventually I would also like to transfer the ROIs to a third channel image for quantification of a third color. And ultimately I would like to automate this to batch process hundreds of images like this. Is there a way to use “Vonoroi” or “Find Maxima” in such a way that the edges of the ROIs end with the edges of the cytoplasmic signal (Actinin)? Or is there a different way to obtain these ROIs? Thanks again, Michiel <http://imagej.1557.n6.nabble.com/file/n5001115/CytoplasmWithROI.jpg> <http://imagej.1557.n6.nabble.com/file/n5001115/NucleiWithROI.jpg> -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001115.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Michiel van Dommelen
Hi Michiel,
I think you could do a threshold on the cytoplasma signal and use the results from the Vonoroi or Find Maxima to separate attached cytoplasm. After that run again a Analyze Particles to get the ROI for the cytoplasm. If you could post two images without ROIs I could test it tomorrow. Best wishes Kees -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: 11 December 2012 14:30 To: [hidden email] Subject: Re: object-oriented segmentation Dear Kees, Thanks for the quick response. I am afraid this is not exactly what I am looking for. The way I interpreted your two methods, both “Vonoroi” as well as “Find Maxima” on the nuclear signal do not take the cytoplasmic signal into account. This way, the ROIs that are created are much bigger than the cytoplasmic signal. Or am I misinterpreting your advice? I will be a bit more explanatory in my question. This time I included two images, one with nuclear signal (Hoechst) and one with cytoplasmic signal (Actinin), processed with a different program. The segmentation is not perfect, but this is more or less what I would like to do with ImageJ. Unfortunately not all of the cells have Actinin, on average about 10% of the nuclei do not have Actinin. I am only interested in the cells that do contain Actinin. Two of the parameters I am looking for are cellular area (either pixels or square um) and average intensity per cell. Eventually I would also like to transfer the ROIs to a third channel image for quantification of a third color. And ultimately I would like to automate this to batch process hundreds of images like this. Is there a way to use “Vonoroi” or “Find Maxima” in such a way that the edges of the ROIs end with the edges of the cytoplasmic signal (Actinin)? Or is there a different way to obtain these ROIs? Thanks again, Michiel <http://imagej.1557.n6.nabble.com/file/n5001115/CytoplasmWithROI.jpg> <http://imagej.1557.n6.nabble.com/file/n5001115/NucleiWithROI.jpg> -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001115.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
Hey Kees,
Your new method sounds interesting. Below you can find the images without ROIs. The original images are 12bit TIF files. I had to transform them into 8bit JPEG to upload them here. I am looking forward to see if this works.. Many thanks, Michiel 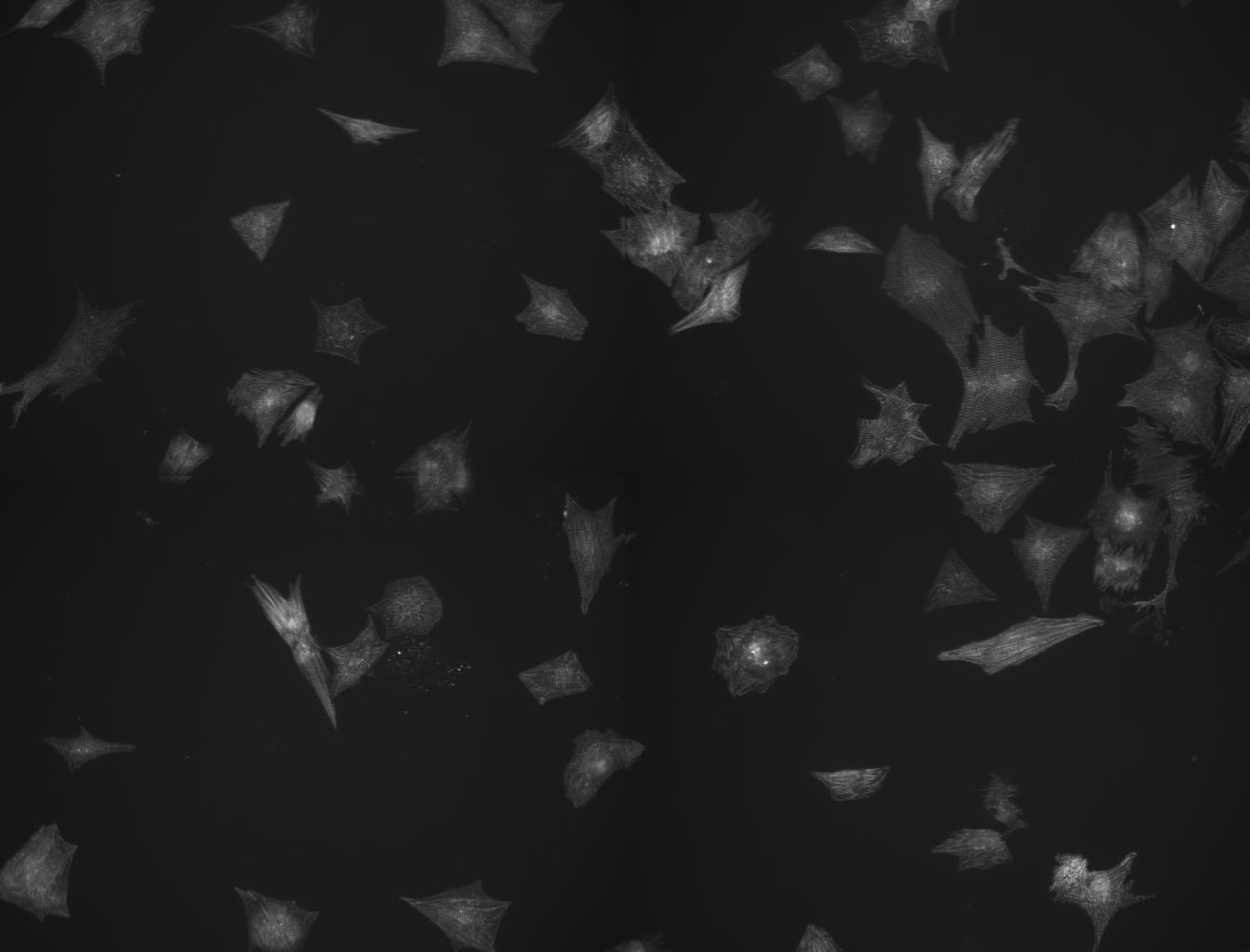 
|
|
Hi Michiel,
The macro code below is doing what you have in mind for the images you sent. You might have play around with some of the settings to have it working for other images. Both images have to be opened when you run the code. There seems to be a problem when I use run("Flatten") in the macro. I get a black/red image (with white ROI lines) instead of a black/white image. If I run it manually it works fine. Did an extra thresholding to solve this problem. selectWindow("Nuclei_Raw.jpg"); setAutoThreshold("Huang dark"); //run("Threshold..."); setAutoThreshold("Huang dark"); setAutoThreshold("Default dark"); setThreshold(34, 255); run("Convert to Mask"); run("Watershed"); run("Voronoi"); setThreshold(0, 0); run("Convert to Mask"); run("Analyze Particles...", "size=0-Infinity circularity=0.00-1.00 show=Nothing add"); roiManager("Show All with labels"); roiManager("Show All"); selectWindow("Cytoplasm_Raw.jpg"); run("Duplicate...", "title=Cytoplasm_Raw-o.jpg"); selectWindow("Cytoplasm_Raw.jpg"); //run("Threshold..."); setAutoThreshold("Default dark"); setThreshold(35, 255); run("Convert to Mask"); roiManager("Show All without labels"); roiManager("Set Color", "white"); roiManager("Set Line Width", 0); run("Flatten"); //at this point should be B/W but selection is in red with white ROIs run("8-bit"); // seems to be a problem with flatten when run in the macro. // Need additonal thresholding setThreshold(71, 100); run("Convert to Mask"); roiManager("Delete");wait(1000); selectWindow("Cytoplasm_Raw-1.jpg"); run("Analyze Particles...", "size=250-Infinity circularity=0.00-1.00 show=Nothing add"); selectWindow("Cytoplasm_Raw-o.jpg"); roiManager("Show All"); Groetjes Kees -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: 12 December 2012 10:50 To: [hidden email] Subject: Re: object-oriented segmentation Hey Kees, Your new method sounds interesting. Below you can find the images without ROIs. The original images are 12bit TIF files. I had to transform them into 8bit JPEG to upload them here. I am looking forward to see if this works.. Many thanks, Michiel <http://imagej.1557.n6.nabble.com/file/n5001130/Cytoplasm_Raw.jpg> <http://imagej.1557.n6.nabble.com/file/n5001130/Nuclei_Raw.jpg> -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001130.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
Hey Kees,
Wauw! Bedankt! This is pretty amazing! Thanks! There is one thing that worries me a bit. I don’t know if this issue can be solved. In the attached image, you will find “Cytoplasm_Raw-1.jpg” in which the segmentation of the cells is visible. In red I have located some of the issues I am talking about. In these cases, a part of the cells is cut off, which results in an underestimation of cell area. Normally it would not be a big problem, but some of my cell treatments result in a shape change of these cells from “more round” to “more elongated”. And especially the elongated cells will be cut off more easily… Would you see a possible solution for this? Thanks, Michiel 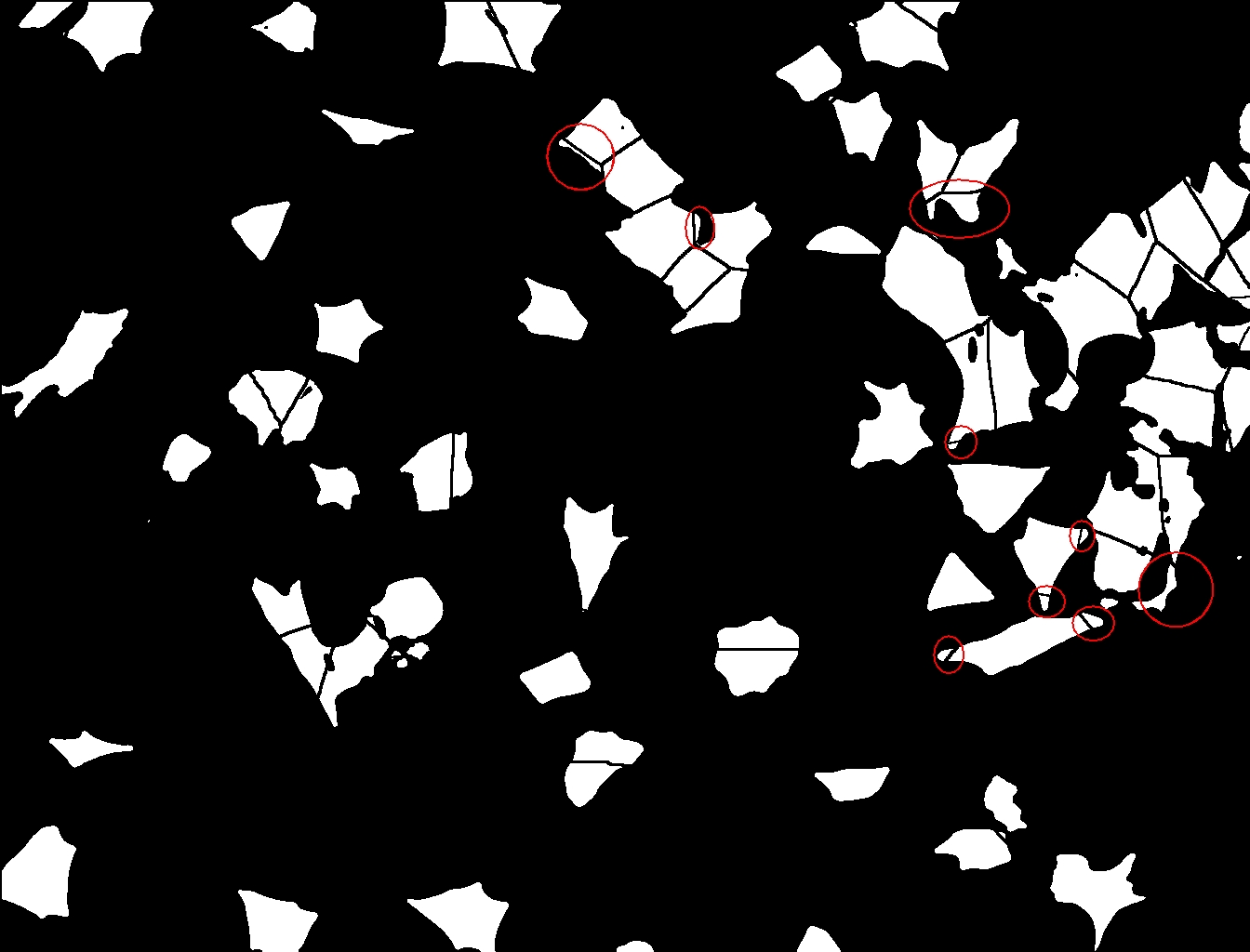
|
Re: object-oriented segmentation
|
In reply to this post by daschneider9
Hi Dave,
I am familiar with JACop. I had a look at this option, but I could not find a way to make use of this. Or would you have an idea on how? Thanks, Michiel |
Re: object-oriented segmentation
|
My apology, Michiel.
As I continued to watch others' replies to your request I realized that I had read your initial email too hurriedly. No, as far as I am aware JACoP is not going to be helpful to your situation. I mistakenly thought you were after object-based colocalization. Again, my apology for making a hasty response. Best wishes, Dave -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: Wednesday, December 12, 2012 6:45 AM To: [hidden email] Subject: Re: object-oriented segmentation Hi Dave, I am familiar with JACop. I had a look at this option, but I could not find a way to make use of this. Or would you have an idea on how? Thanks, Michiel -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001137.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Michiel van Dommelen
Hi Michiel,
Yes that is the result of using the nuclear image to segment the cytoplasmic signal. Not sure 1,2,3 how to improve that. You could try the watershed plugin from Daniel Saga (http://bigwww.epfl.ch/sage/soft/watershed/) or Christopher Mei (http://rsbweb.nih.gov/ij/plugins/watershed.html) to see if they give a better result for this. Best wishes Kees -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of Michiel van Dommelen Sent: 12 December 2012 14:03 To: [hidden email] Subject: Re: object-oriented segmentation Hey Kees, Wauw! Bedankt! This is pretty amazing! Thanks! There is one thing that worries me a bit. I don’t know if this issue can be solved. In the attached image, you will find “Cytoplasm_Raw-1.jpg” in which the segmentation of the cells is visible. In red I have located some of the issues I am talking about. In these cases, a part of the cells is cut off, which results in an underestimation of cell area. Normally it would not be a big problem, but some of my cell treatments result in a shape change of these cells from “more round” to “more elongated”. And especially the elongated cells will be cut off more easily… Would you see a possible solution for this? Thanks, Michiel <http://imagej.1557.n6.nabble.com/file/n5001136/Cytoplasm_Raw-1.jpg> -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001136.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
Hey Kees,
Thanks! Your analysis protocol is already so much better than I had found out by myself. I will have a look at these watershed options! We'll see if these can give some improvements! Warm regards, Michiel 2012/12/12 Straatman, Kees R. (Dr.) <[hidden email]> > Hi Michiel, > > Yes that is the result of using the nuclear image to segment the > cytoplasmic signal. Not sure 1,2,3 how to improve that. You could try the > watershed plugin from Daniel Saga ( > http://bigwww.epfl.ch/sage/soft/watershed/) or Christopher Mei ( > http://rsbweb.nih.gov/ij/plugins/watershed.html) to see if they give a > better result for this. > > Best wishes > > Kees > > -----Original Message----- > From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of > Michiel van Dommelen > Sent: 12 December 2012 14:03 > To: [hidden email] > Subject: Re: object-oriented segmentation > > Hey Kees, > > Wauw! Bedankt! This is pretty amazing! Thanks! > There is one thing that worries me a bit. I don’t know if this issue can > be solved. In the attached image, you will find “Cytoplasm_Raw-1.jpg” in > which the segmentation of the cells is visible. In red I have located some > of the issues I am talking about. In these cases, a part of the cells is > cut off, which results in an underestimation of cell area. Normally it > would not be a big problem, but some of my cell treatments result in a > shape change of these cells from “more round” to “more elongated”. And > especially the elongated cells will be cut off more easily… Would you see a > possible solution for this? > > Thanks, > Michiel > <http://imagej.1557.n6.nabble.com/file/n5001136/Cytoplasm_Raw-1.jpg> > > > > -- > View this message in context: > http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001136.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
In reply to this post by Krs5
Hey Kees,
I got it working! I used the plugin from Daniel Saga (http://bigwww.epfl.ch/sage/soft/watershed/). The only thing I do not understand is why the "exclude on edge" functionality is not working in this code. Would you have any idea? Of course I have to tweak some settings to make it robust enough for higher throughput. For the rest it is working like a charm. I got it working with the code below: selectWindow("Cytoplasm_Raw.jpg"); run("Duplicate...", "title=[Cytoplasm_Raw_temp.jpg]"); run("Median...", "radius=5"); run("Auto Threshold", "method=Triangle white"); run("Divide...", "value=2"); //this makes the cytoplasm grey in dual color overlay selectWindow("Nuclei_Raw.jpg"); run("Duplicate...", "title=[Nuclei_Raw_temp.jpg]"); run("Median...", "radius=5"); run("Auto Threshold", "method=Triangle white"); run("Watershed"); imageCalculator("Add", "Nuclei_Raw_temp.jpg","Cytoplasm_Raw_temp.jpg"); //this makes dual color overlay rename("2ColorOverlay"); selectWindow("Cytoplasm_Raw_temp.jpg"); close(); selectWindow("2ColorOverlay"); run("Watershed ", "blurring='0' watershed='1 1 0 100 0 0' display='0' "); //this is the plugin I installed selectWindow("Binary watershed lines"); run("Invert LUT"); run("Set Scale...", "distance=1 known=0.617 pixel=1 unit=um global"); //These 2 lines adjust the settings specific to my images run("Analyze Particles...", "size=350-Infinity circularity=0.00-1.00 show=Nothing exclude clear include add");//These 2 lines adjust the settings specific to my images selectWindow("Nuclei_Raw.jpg"); roiManager("Show All with labels"); roiManager("Show All"); selectWindow("Cytoplasm_Raw.jpg"); roiManager("Show All with labels"); roiManager("Show All"); Thanks for all your help! Warm regards, Michiel |
|
Hi Michiel,
It looks like the plugin Watershed makes the pixels at the boarder white with as result that the cells are not recognized as edge anymore. If you add makeRectangle(2, 1, 1341, 1021); before you run Analyse Particles it should go OK. Best wishes Kees ________________________________________ From: ImageJ Interest Group [[hidden email]] On Behalf Of Michiel van Dommelen [[hidden email]] Sent: 14 December 2012 11:51 To: [hidden email] Subject: Re: object-oriented segmentation Hey Kees, I got it working! I used the plugin from Daniel Saga ( http://bigwww.epfl.ch/sage/soft/watershed/ <http://bigwww.epfl.ch/sage/soft/watershed/> ). The only thing I do not understand is why the "exclude on edge" functionality is not working in this code. Would you have any idea? Of course I have to tweak some settings to make it robust enough for higher throughput. For the rest it is working like a charm. I got it working with the code below: selectWindow("Cytoplasm_Raw.jpg"); run("Duplicate...", "title=[Cytoplasm_Raw_temp.jpg]"); run("Median...", "radius=5"); run("Auto Threshold", "method=Triangle white"); run("Divide...", "value=2"); //this makes the cytoplasm grey in dual color overlay selectWindow("Nuclei_Raw.jpg"); run("Duplicate...", "title=[Nuclei_Raw_temp.jpg]"); run("Median...", "radius=5"); run("Auto Threshold", "method=Triangle white"); run("Watershed"); imageCalculator("Add", "Nuclei_Raw_temp.jpg","Cytoplasm_Raw_temp.jpg"); //this makes dual color overlay rename("2ColorOverlay"); selectWindow("Cytoplasm_Raw_temp.jpg"); close(); selectWindow("2ColorOverlay"); run("Watershed ", "blurring='0' watershed='1 1 0 100 0 0' display='0' "); //this is the plugin I installed selectWindow("Binary watershed lines"); run("Invert LUT"); run("Set Scale...", "distance=1 known=0.617 pixel=1 unit=um global"); //These 2 lines adjust the settings specific to my images run("Analyze Particles...", "size=350-Infinity circularity=0.00-1.00 show=Nothing exclude clear include add");//These 2 lines adjust the settings specific to my images selectWindow("Nuclei_Raw.jpg"); roiManager("Show All with labels"); roiManager("Show All"); selectWindow("Cytoplasm_Raw.jpg"); roiManager("Show All with labels"); roiManager("Show All"); Thanks for all your help! Warm regards, Michiel -- View this message in context: http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001161.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
Re: object-oriented segmentation
|
Hey Kees,
That works perfectly! Many thanks, Michiel 2012/12/15 Straatman, Kees R. (Dr.) <[hidden email]> > Hi Michiel, > > It looks like the plugin Watershed makes the pixels at the boarder white > with as result that the cells are not recognized as edge anymore. If you add > > makeRectangle(2, 1, 1341, 1021); > > before you run Analyse Particles it should go OK. > > Best wishes > > Kees > ________________________________________ > From: ImageJ Interest Group [[hidden email]] On Behalf Of Michiel > van Dommelen [[hidden email]] > Sent: 14 December 2012 11:51 > To: [hidden email] > Subject: Re: object-oriented segmentation > > Hey Kees, > > I got it working! I used the plugin from Daniel Saga ( > http://bigwww.epfl.ch/sage/soft/watershed/ > <http://bigwww.epfl.ch/sage/soft/watershed/> ). The only thing I do not > understand is why the "exclude on edge" functionality is not working in > this > code. Would you have any idea? Of course I have to tweak some settings to > make it robust enough for higher throughput. For the rest it is working > like > a charm. > > I got it working with the code below: > > selectWindow("Cytoplasm_Raw.jpg"); > run("Duplicate...", "title=[Cytoplasm_Raw_temp.jpg]"); > run("Median...", "radius=5"); > run("Auto Threshold", "method=Triangle white"); > run("Divide...", "value=2"); //this makes the cytoplasm grey in dual color > overlay > selectWindow("Nuclei_Raw.jpg"); > run("Duplicate...", "title=[Nuclei_Raw_temp.jpg]"); > run("Median...", "radius=5"); > run("Auto Threshold", "method=Triangle white"); > run("Watershed"); > imageCalculator("Add", "Nuclei_Raw_temp.jpg","Cytoplasm_Raw_temp.jpg"); > //this makes dual color overlay > rename("2ColorOverlay"); > selectWindow("Cytoplasm_Raw_temp.jpg"); > close(); > selectWindow("2ColorOverlay"); > run("Watershed ", "blurring='0' watershed='1 1 0 100 0 0' display='0' > "); //this is the plugin I installed > selectWindow("Binary watershed lines"); > run("Invert LUT"); > run("Set Scale...", "distance=1 known=0.617 pixel=1 unit=um global"); > //These 2 lines adjust the settings specific to my images > run("Analyze Particles...", "size=350-Infinity circularity=0.00-1.00 > show=Nothing exclude clear include add");//These 2 lines adjust the > settings > specific to my images > selectWindow("Nuclei_Raw.jpg"); > roiManager("Show All with labels"); > roiManager("Show All"); > selectWindow("Cytoplasm_Raw.jpg"); > roiManager("Show All with labels"); > roiManager("Show All"); > > Thanks for all your help! > Warm regards, > Michiel > > > > -- > View this message in context: > http://imagej.1557.n6.nabble.com/object-oriented-segmentation-tp5001110p5001161.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

