optical coherence tomography (OCT) segmentation - ganglion cell layer thickness
|
Hi all,
Some of you might be familiar with the following OCT image of a rodent eye:  Due to the hyper-reflectivity of the ganglion cell layer (GCL), Ive found a quick way to segment it out using thresholding. 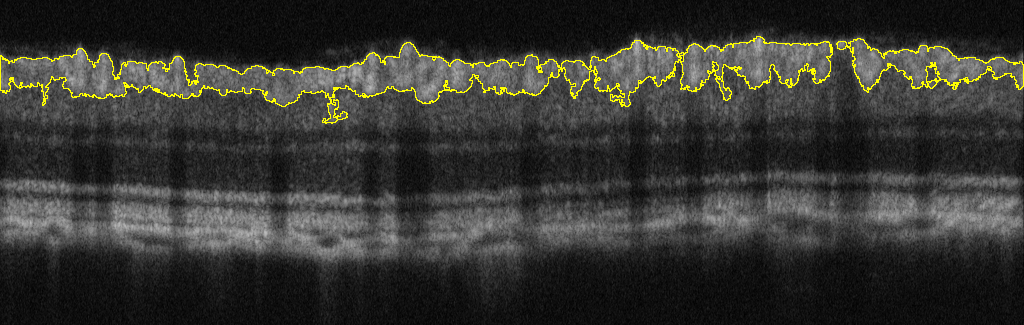 My question is, now that I have made a composite selection of the GCL, is there a way to find out the thickness of this ROI? I'm looking to get the average thickness and maximum thickness of the segmented layer. 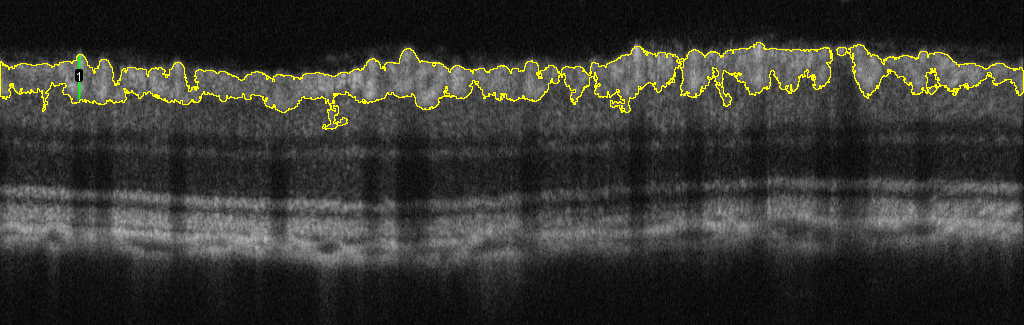 Ideally, I would like a measurement of the height/length of the 1-pixel wide green line (see example above) throughout the entire length of the ROI (1024 points) so that I can average all of them (which also gives me the max height). Any help would be appreciated. Thank you. |
Re: optical coherence tomography (OCT) segmentation - ganglion cell layer thickness
|
Hi,
You may want to use the Local Thickness plugin : http://www.optinav.com/imagej.html I think it is available in Fij. Best, Thomas > Hi all, > > Some of you might be familiar with the following OCT image of a rodent > eye: > <http://imagej.1557.x6.nabble.com/file/n5009735/OCT_pre_segmented.png> > > Due to the hyper-reflectivity of the ganglion cell layer (GCL), Ive found > a > quick way to segment it out using thresholding. > <http://imagej.1557.x6.nabble.com/file/n5009735/OCT_segmented_GCL.png> > > My question is, now that I have made a composite selection of the GCL, is > there a way to find out the thickness of this ROI? I'm looking to get the > average thickness and maximum thickness of the segmented layer. > > <http://imagej.1557.x6.nabble.com/file/n5009735/OCT_segmented_GCL_height.png> > > Ideally, I would like a measurement of the height/length of the 1-pixel > wide > green line (see example above) throughout the entire length of the ROI > (1024 > points) so that I can average all of them (which also gives me the max > height). > > Any help would be appreciated. Thank you. > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/optical-coherence-tomography-OCT-segmentation-ganglion-cell-layer-thickness-tp5009735.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > -- Thomas Boudier, Associate Prof. UPMC, IPAL UMI 2955, Singapore. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Thanks Thomas for your prompt response. Ive tried the plugin and and I got this:
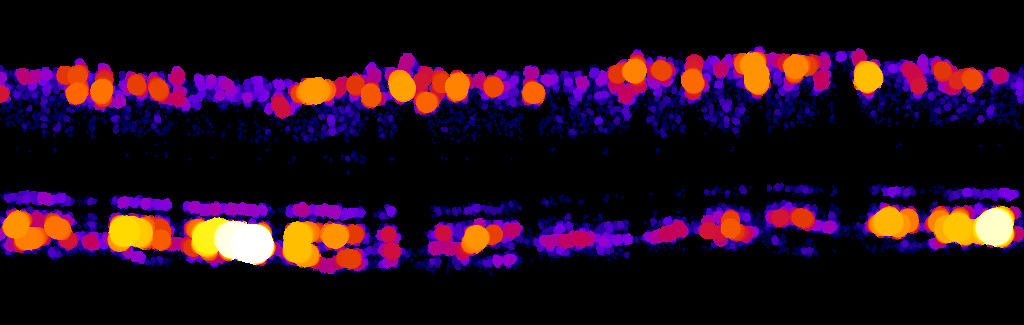 However, I'm not sure how to extract the data i.e. how do I get the individual diameter values for each circle? Is there a way to spit out the values for analysis in excel? |
Re: optical coherence tomography (OCT) segmentation - ganglion cell layer thickness
|
Dear Mr_Sonky
It would appear you are not using the local thickness plugin properly. If you managed to make a selection based on simple thresholding, you should apply the local thickness on your thresholded image. Or you could convert your selection to a mask and then apply the local thickness plugin. From the image you submitted, it seems you simply set another threshold on the original image when launching the local thickness plugin which gave the fragmented local thickness map that you have. You also seem to be only interested in the thickness of the cell layer in the vertical direction. Local thickness will also give you information on the small protrusions where the local thickness is influenced by other directions, so I am not sure if it is that useful to you. Then I might suggest the following approach. After you have got your selection just the way you want it, run the following macro code: run("Clear Results"); run("Set Measurements...", " bounding stack limit display redirect=None decimal=3"); run("To Bounding Box"); run("Reslice [/]...", "output=1.000 start=Left avoid"); run("Analyze Particles...", "display clear stack"); // Check slice number getDimensions(x,y,c,z,t); theWidth = newArray(z); name=getTitle(); for (i=0;i<nResults;i++) { tmpW = getResult("Width", i); slice = getResult("Slice", i); theWidth[slice-1]+=tmpW; } run("Clear Results"); for (i=0;i<z;i++) { setResult("Label", i, name); setResult("Position", i, i+1); setResult("Thickness", i, theWidth[i]); } What this does is to create a box around your selection and reslicing the box along the y direction. What you get at the end is a stack of 1D images that basically represent your mask being swept by a vertical line from left to right. Then we run analyze particles on that stack of images to measure the length of each segment, which is what you want in the end. Finally, because of the shape of your mask, you might get some dissociated particles. I have chosen to add the values so that you have one thickness per line only. Feel free to modify this as you see fit. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
This post was updated on .
Dear Olivier,
I've tried your macro and its works beautifully!! It does exactly what I had in mind. I really cant thank you enough   Thank you so much for your time and the effort put into writing this script for me :) |
|
In reply to this post by Mr_Sonky
Hello,
I also work with OCT's and we are trying to transition to ImageJ to grade the layers. Could you explain how you were able to section out the layers using the Threshold function? Thank you |
|
Mascuit, I find that this method works only when the OCT signal is really good or if the image is very clean. I ended up using manual segmentation for my own study. However, i created a screencast (using imageJ) of me doing the segmentation. The video (click link below) may be played using VLC player.
OCT_segmentation_video.ogv |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

