Nuclei segmentation
|
Greetings,
Are there better ways to segment those various types of nuclei in a image? I tried some threshold methods to segment then use watershed to split connect nuclei. But it seems not be segmented completely by using those methods. Please someone could try it and give me some advice. I appreciate any suggestion. chin 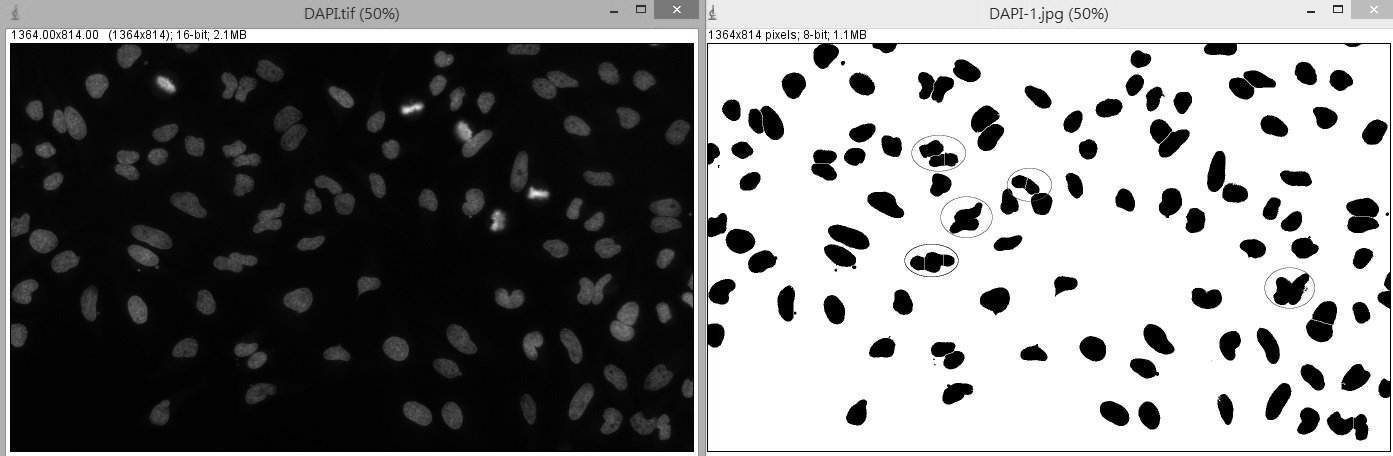 The original image: http://postimg.org/image/alba8j6u1/ |
|
Hi Chin,
On Mon, 17 Mar 2014, chin wrote: > Are there better ways to segment those various types of nuclei in a image? > I tried some threshold methods to segment then use watershed to split > connect nuclei. But it seems not be segmented completely by using those > methods. You might need to apply some background subtraction with a real large diameter for the threshold, and then use the "Watershed Irregular Structures" command in the BioVoxxel Toolbox: http://fiji.sc/BioVoxxel_Toolbox#Watershed_Irregular_Structures Ciao, Johannes -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
What about supervised segmentation with weka or ilastik?
On Mar 17, 2014 11:57 AM, "Johannes Schindelin" <[hidden email]> wrote: > Hi Chin, > > On Mon, 17 Mar 2014, chin wrote: > > > Are there better ways to segment those various types of nuclei in a > image? > > I tried some threshold methods to segment then use watershed to split > > connect nuclei. But it seems not be segmented completely by using those > > methods. > > You might need to apply some background subtraction with a real large > diameter for the threshold, and then use the "Watershed Irregular > Structures" command in the BioVoxxel Toolbox: > > http://fiji.sc/BioVoxxel_Toolbox#Watershed_Irregular_Structures > > Ciao, > Johannes > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Those tools are both great as long as input size isn't too large!
On Mar 17, 2014 12:03 PM, "Adam Hughes" <[hidden email]> wrote: > What about supervised segmentation with weka or ilastik? > On Mar 17, 2014 11:57 AM, "Johannes Schindelin" < > [hidden email]> > wrote: > > > Hi Chin, > > > > On Mon, 17 Mar 2014, chin wrote: > > > > > Are there better ways to segment those various types of nuclei in a > > image? > > > I tried some threshold methods to segment then use watershed to split > > > connect nuclei. But it seems not be segmented completely by using those > > > methods. > > > > You might need to apply some background subtraction with a real large > > diameter for the threshold, and then use the "Watershed Irregular > > Structures" command in the BioVoxxel Toolbox: > > > > http://fiji.sc/BioVoxxel_Toolbox#Watershed_Irregular_Structures > > > > Ciao, > > Johannes > > > > -- > > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > > > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by dscho
Hi Chin, Johannes, William and Adam,
In such a case the watershed irregular features will unfortunately hit its limit. It does the job as long as the connections between features are "thin" compared to the features themselfes and to further protrusions. This is unfortunately not the case for those nuclei. Besides the fact, that I need to improve its performance I was also thinking already about additional methods which might lead to a seperation of features where the normal watershed fails. So far, no good method came to my mind to achieve this. So, I am happy about any suggestions and would try to include them in the toolbox. cheers, Jan 2014-03-17 16:56 GMT+01:00 Johannes Schindelin <[hidden email]>: > Hi Chin, > > On Mon, 17 Mar 2014, chin wrote: > > > Are there better ways to segment those various types of nuclei in a > image? > > I tried some threshold methods to segment then use watershed to split > > connect nuclei. But it seems not be segmented completely by using those > > methods. > > You might need to apply some background subtraction with a real large > diameter for the threshold, and then use the "Watershed Irregular > Structures" command in the BioVoxxel Toolbox: > > http://fiji.sc/BioVoxxel_Toolbox#Watershed_Irregular_Structures > > Ciao, > Johannes > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
This post was updated on .
Thank you all your answer.
Yeah, it seems no better ways to use watershed method to split connections between such irregular objects right now. I could only do is draw the lines to split them manully. But it takes much time and quite laborious job. Hopefully, there will be some labor-saving method for imagej developed by some experts. I tried the "Squassh" plugin of "MosaicSuite for ImageJ and Fiji to segment nuclei. It could draw outlines of nuclei, though, the connection of objects still could not be split... best, chin |
|
Hi Chin,
watershed (more exactly, watershed of the Euclidian Distance map) does not help because it is essentially meant for circular or near-circular particles. The only chance for better segmentation I see is trying to enhance the weak dark lines where the nuclei touch, and make them dividing lines in the binary image. Then watershed might do the job. Michael ________________________________________________________________ On Mar 18, 2014, at 08:14, chin wrote: > Thank you all your answer. > > Yeah, it seems no better ways to use watershed method to split connections > between such irregular > objects right now. I could only do is draw the lines to split them manully. > But it takes much time and > quite laborious job. Hopefully, there will be some labor-saving method for > imagej developed by some > experts. > > best, > chin > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Nuclei-segmentation-tp5006953p5006964.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
Hi Michael,
Thank you for the suggestion. I tried to enhance the contrast then did the segmentation. The separation of connective objects is a little bit improvment, though, those irregular objects are still in the wrong split position after performing the watershed. best, chin |
|
Hi Chin,
you can try a 'weaker' watershed to avoid oversegmentation. I have uploaded the Adjustable Watershed to the documentation wiki, there you can try with the 'tolerance' parameter. http://imagejdocu.tudor.lu/doku.php?id=plugin:segmentation:adjustable_watershed:start Nevertheless, watershed of the Euclidian Distance Map (EDM) works best on roughly circular particles. For elongated particles, one should have different metrics than the EDM, e.g. the two largest radii in opposite directions from any point. If anyone has a plugin that does this instead of the EDM, it would certainly help. Michael ________________________________________________________________ On Mar 19, 2014, at 09:54, chin wrote: > Hi Michael, > > Thank you for the suggestion. I tried to enhance the contrast then did the > segmentation. The separation > of connective objects is a little bit improvment, though, those irregular > objects are still in the wrong split > position after performing the watershed. > > best, > chin > > > > -- > View this message in context: http://imagej.1557.x6.nabble.com/Nuclei-segmentation-tp5006953p5006985.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by chin
Hi Chin,
I have written some macro code that might be useful. You probably have to change some of the settings but this works on the image you posted. It also uses the adjustable Watershed plugin as posted by Michael. It will threshold an open image, run the adjustable watershed, get the perimeter values and all particles with a perimeter above 220 are extracted one by one to allow you to manually separate them. You might have to change this value in case too many wrong cells are selected. -------------------------------- start code-------------------------- waitForUser("Please select the pencil tool"); run("Set Measurements...", " perimeter redirect=None decimal=3"); setForegroundColor(255, 255, 255); if (isOpen("ROI Manager")) { selectWindow("ROI Manager"); run("Close"); } title = getTitle(); setAutoThreshold("Huang dark"); setOption("BlackBackground", false); run("Convert to Mask"); run("Adjustable Watershed", "tolerance=2"); run("Analyze Particles...", "display add"); roiManager("Deselect"); for (i=0;i<roiManager("count");i++){ roiManager("select", i) run("Measure"); if (getResult("Perim.", i)>220){ // You might have to change this number run("To Bounding Box"); run("Enlarge...", "enlarge=1"); Roi.getBounds(x, y, width, height); run("Duplicate...", "title=test"); run("Set... ", "zoom=200"); // You can change the zoom waitForUser("Separate Cells"); run("Copy"); selectWindow(title); makeRectangle(x, y, width, height); run("Paste"); selectWindow("test"); close(); } } roiManager("Show None"); selectWindow("ROI Manager"); run("Close"); selectWindow("Results"); run("Close"); -----end----- Best wishes Kees Dr Ir K.R. Straatman Senior Experimental Officer Centre for Core Biotechnology Services University of Leicester http://www2.le.ac.uk/colleges/medbiopsych/facilities-and-services/cbs/lite/aif ImageJ workshops 14 and 15 April: http://www2.le.ac.uk/colleges/medbiopsych/facilities-and-services/cbs/lite/aif/workshops/imagej-workshops-14-and-15-April-2014 (Still some places available) -----Original Message----- From: ImageJ Interest Group [mailto:[hidden email]] On Behalf Of chin Sent: 18 March 2014 07:14 To: [hidden email] Subject: Re: Nuclei segmentation Thank you all your answer. Yeah, it seems no better ways to use watershed method to split connections between such irregular objects right now. I could only do is draw the lines to split them manully. But it takes much time and quite laborious job. Hopefully, there will be some labor-saving method for imagej developed by some experts. best, chin -- View this message in context: http://imagej.1557.x6.nabble.com/Nuclei-segmentation-tp5006953p5006964.html Sent from the ImageJ mailing list archive at Nabble.com. -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
|
In reply to this post by Michael Schmid
Hi Michael,
Thank you for the Adjustable Watershed. It does pretty well on segmentation when some non-circular objects need not to be segmented. I tried the "Tolerance" of 2.3 and obtained the satisfied segmentation. There are only four pairs of nuclei contact side by side. This plugin is really useful not only for me but for those who want to use more flexible watershed to segment their objects. ----------------------- Hi Kees, Your macro gives me another approach to use Michael's plugin. I didn't mention about the unit of original image is um. The macro didn't show the "Action Required: Separate cells" by using the perim>220. So I change the value to 90, the result looks great. Thank you for your assistance. Again, Thank both Michael and Kees. best, chin |
|
Hi all,
I also tried the adjustable watershed plugin which is really good and I think it would be a good idea to integrate this in one of the following new versions of ImageJ or distribute it by default with the Fiji package. Very nice and useful tool (actually makes the 'watershed irregular features' macro from the biovoxxel toolbox kind of obsolete, but I still haven't compared the performances of both in different images). cheers, Jan 2014-03-20 7:28 GMT+01:00 chin <[hidden email]>: > Hi Michael, > > Thank you for the Adjustable Watershed. It does pretty well on segmentation > when some non-circular > objects need not to be segmented. I tried the "Tolerance" of 2.3 and > obtained the satisfied segmentation. > There are only four pairs of nuclei contact side by side. > > This plugin is really useful not only for me but for those who want to use > more flexible watershed to > segment their objects. > > ----------------------- > > Hi Kees, > > Your macro gives me another approach to use Michael's plugin. I didn't > mention about the unit of original > image is um. The macro didn't show the "Action Required: Separate cells" by > using the perim>220. So I > change the value to 90, the result looks great. Thank you for your > assistance. > > Again, Thank both Michael and Kees. > > best, > chin > > > > > > -- > View this message in context: > http://imagej.1557.x6.nabble.com/Nuclei-segmentation-tp5006953p5007007.html > Sent from the ImageJ mailing list archive at Nabble.com. > > -- > ImageJ mailing list: http://imagej.nih.gov/ij/list.html > -- CEO: Dr. rer. nat. Jan Brocher phone: +49 (0)6234 917 03 39 mobile: +49 (0)176 705 746 81 e-mail: [hidden email] info: [hidden email] inquiries: [hidden email] web: www.biovoxxel.de -- ImageJ mailing list: http://imagej.nih.gov/ij/list.html |
«
Return to ImageJ
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

